BAND fragment job¶
In this module a dedicated job type for Energy Decomposition Analysis in BAND is defined.
Such an analysis is performed on a periodic system divided into 2 fragments and consists of a minimum of 3 separate BAND runs: one for each fragment and one for full system. See also ADFFragmentJob.
We define a new job type BANDFragmentJob, by subclassing ADFFragmentJob, which in turn is a subclass of MultiJob.
The constructor (__init__) of this new job takes 2 more arguments (fragment1 and fragment2) and one optional argument full_settings for additional input keywords that are used only in the full system calculation. Furthermore, you can specify
the optimized fragment geometries using fragment1_opt and fragment2_opt for single-point calculations to also obtain the preparation energies.
In the prerun() method two fragment jobs and the full system job are created with the proper settings and molecules.
They are then added to the children list.
The dedicated Results subclass for BANDFragmentJob does not provide too much additional functionality.
It simply redirects the usual AMSResults methods to the results of the full system calculation. The pEDA results can be obtained using the get_energy_decomposition method. It will return a dictionary with the available energy decomposition terms.
A derived subclass NOCVBandFragmentJob is also provided. It can be usefull for generating NOCV plots after the PEDA-NOCV calculation.
from scm.plams.core.functions import log
from scm.plams.interfaces.adfsuite.ams import AMSJob
from scm.plams.core.settings import Settings
from scm.plams.recipes.adffragment import ADFFragmentJob, ADFFragmentResults
from scm.plams.tools.units import Units
from scm.plams.core.errors import FileError
from scm.plams.core.functions import add_to_instance
from scm.plams.core.enums import JobStatus
from os.path import join as opj
from os.path import abspath, isdir, basename, relpath
from os import symlink
from typing import Dict, List, Union, Optional
__all__ = ["BANDFragmentJob", "BANDFragmentResults"]
class BANDFragmentResults(ADFFragmentResults):
"""Subclass of |ADFFragmentResults| for BAND calculations."""
def get_energy_decomposition(self, unit="au") -> Dict[str, float]:
"""Get the energy decomposition of the fragment calculation.
Args:
unit (str, optional): The unit of the energy. Defaults to 'au'.
Returns:
Dict[str, float]: The energy decomposition.
"""
res = self.job.full.results
res1 = self.job.f1.results
res2 = self.job.f2.results
ret = {}
def try_grep_result(key, pattern, match_position=-1, split_position=2):
match = res.grep_output(pattern)
if match:
match = Units.convert(float(match[match_position].split()[split_position]), "au", unit)
ret[key] = match
try_grep_result("E_int", "E_int", match_position=-2)
try_grep_result("E_int_disp", "E_disp")
try_grep_result("E_Pauli", "E_Pauli")
try_grep_result("E_elstat", "E_elstat")
try_grep_result("E_orb", "E_orb")
ret["E_1"] = res1.get_energy(unit=unit)
ret["E_2"] = res2.get_energy(unit=unit)
if hasattr(self.job, "f1_opt"):
ret["E_1_opt"] = self.job.f1_opt.results.get_energy(unit=unit)
ret["E_prep_1"] = ret["E_1"] - ret["E_1_opt"]
if hasattr(self.job, "f2_opt"):
ret["E_2_opt"] = self.job.f2_opt.results.get_energy(unit=unit)
ret["E_prep_2"] = ret["E_2"] - ret["E_2_opt"]
if ("E_1_opt" in ret) and ("E_2_opt" in ret):
ret["E_prep"] = ret["E_prep_1"] + ret["E_prep_2"]
ret["E_bond"] = ret["E_int"] + ret["E_prep"]
return ret
class BANDFragmentJob(ADFFragmentJob):
_result_type = BANDFragmentResults
def prerun(self) -> None:
"""Creates the fragment jobs."""
self.f1 = AMSJob(name="frag1", molecule=self.fragment1, settings=self.settings)
self.f2 = AMSJob(name="frag2", molecule=self.fragment2, settings=self.settings)
self.children += [self.f1, self.f2]
def new_children(self) -> Union[None, List[AMSJob]]:
"""After the first round, add the full job to the children list."""
if hasattr(self, "full"):
return None
else:
# create the correct mapping settings for the full job
if "fragment" in self.full_settings.input.band:
log(
"Fragment already present in full_settings. Assuming that the user has already set up the mapping. Skipping the mapping setup.",
level=1,
)
return
# first fragment 1 then fragment 2
set1 = Settings()
set1.atommapping = {str(i + 1): str(i + 1) for i in range(len(self.fragment1))}
set2 = Settings()
set2.atommapping = {str(i + 1): str(i + 1 + len(self.fragment1)) for i in range(len(self.fragment2))}
set1.filename = (self.f1, "band")
set2.filename = (self.f2, "band")
self.full_settings.input.band.fragment = [set1, set2]
# create the full job
self.full = AMSJob(
name="full", molecule=self.fragment1 + self.fragment2, settings=self.settings + self.full_settings
)
return [self.full]
@classmethod
def load_external(cls, path: str, jobname: Optional[str] = None) -> "BANDFragmentJob":
"""Load the results of the BANDFragmentJob job from an external path.
Args:
path (str): The path to the job. It should at least have the
subfolders 'frag1', 'frag2' and 'full'.
jobname (str, optional): The name of the job. Defaults to None.
Returns:
BANDFragmentJob: The job with the loaded results.
"""
if not isdir(path):
raise FileError("Path {} does not exist, cannot load from it.".format(path))
path = abspath(path)
jobname = basename(path) if jobname is None else str(jobname)
job = cls(name=jobname)
job.path = path
job.status = JobStatus.COPIED
job.f1 = AMSJob.load_external(opj(path, "frag1"))
job.f2 = AMSJob.load_external(opj(path, "frag2"))
job.full = AMSJob.load_external(opj(path, "full"))
job.children = [job.f1, job.f2, job.full]
if isdir(opj(path, "frag1_opt")):
job.f1_opt = AMSJob.load_external(opj(path, "frag1_opt"))
job.children.append(job.f1_opt)
if isdir(opj(path, "frag2_opt")):
job.f2_opt = AMSJob.load_external(opj(path, "frag2_opt"))
job.children.append(job.f2_opt)
return job
class NOCVBandFragmentJob(BANDFragmentJob):
_result_type = BANDFragmentResults
def __init__(self, nocv_settings=None, **kwargs) -> None:
"""Create a BANDFragmentJob with final NOCV calculation.
Args:
nocv_settings (Settings, optional): Settings for the NOCV calculation. Defaults to None.
"""
super().__init__(**kwargs)
self.nocv_settings = nocv_settings or Settings()
def new_children(self) -> Union[None, List[AMSJob]]:
"""After the first round, add the full job to the children list.
After the second round, add the NOCV job to the children list."""
# new_children of BANDFragmentJob creates the full job
ret = super().new_children()
if ret is not None:
return ret
if hasattr(self, "nocv"):
return None
else:
# add NOCV run
# settings for the NOCV calculation
set = self.settings + self.full_settings + self.nocv_settings
# set the restart file
set.input.band.restart.file = "full.rkf"
# copy the fragment settings
set.input.band.fragment = [fset.copy() for fset in self.full.settings.input.band.fragment]
self.nocv = AMSJob(name="NOCV", molecule=self.fragment1 + self.fragment2, settings=set)
self.nocv.frag_paths = []
for job in [self.f1, self.f2, self.full]:
self.nocv.frag_paths.append(job.path)
# edit NOCV prerun to create symlinks
@add_to_instance(self.nocv)
def prerun(self):
"""Create symlinks for the restart files."""
for i, job in enumerate(["frag1", "frag2", "full"]):
rel_path = relpath(self.frag_paths[i], self.path)
symlink(opj(rel_path, "band.rkf"), opj(self.path, f"{job}.rkf"))
return [self.nocv]
@classmethod
def load_external(cls, path: str, jobname: Optional[str] = None) -> "NOCVBandFragmentJob":
"""Load the results of the BANDFragmentJob job from an external path.
Args:
path (str): The path to the job. It should at least have the
subfolders 'frag1', 'frag2', 'full' and 'NOCV'.
jobname (str, optional): The name of the job. Defaults to None.
Returns:
NOCVBandFragmentJob: The job with the loaded results.
"""
job = super().load_external(path, jobname)
job.nocv = AMSJob.load_external(opj(path, "NOCV"))
job.children.append(job.nocv)
return job
To follow along, either
Download
bandfrag.py(run as$AMSBIN/amspython bandfrag.py).Download
bandfrag.ipynb(see also: how to install Jupyterlab in AMS)
Worked Example¶
Initialization¶
from scm.plams import Settings, fromASE, plot_molecule
from scm.plams.recipes.bandfragment import BANDFragmentJob
# build the surface
from ase import Atoms
from ase.build import fcc111, add_adsorbate
Build Surface & Fragments¶
We first build a gold surface and add the hydrogen adsorbate.
mol = fcc111("Au", size=(2, 2, 3))
add_adsorbate(mol, "H", 1.5, "ontop")
mol.center(vacuum=10.0, axis=2)
plot_molecule(mol);
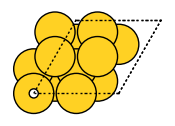
The system is then split into two fragments based on the species.
surface = mol.copy()
symbols = surface.get_chemical_symbols()
del surface[[i for i in range(len(symbols)) if symbols[i] != "Au"]]
adsorbate = mol.copy()
del adsorbate[[i for i in range(len(symbols)) if symbols[i] == "Au"]]
If available, optimized fragments can also be loaded.
# from ase import io
# surface_opt = io.read("surface_opt.xyz")
# adsorbate_opt = io.read("adsorbate_opt.xyz")
# assert len(surface_opt) == len(surface)
# assert len(adsorbate_opt) == len(adsorbate)
Set Up & Run Job¶
For efficiency in this example, we use a minimal basis and reduced computational details to run the job.
base_settings = Settings()
base_settings.input.ams.task = "SinglePoint"
base_settings.input.band.basis.type = "SZ"
base_settings.input.band.basis.core = "Large"
base_settings.input.band.dos.calcdos = "No"
base_settings.input.band.kspace.regular.numberofpoints = "3 3"
base_settings.input.band.beckegrid.quality = "Basic"
base_settings.input.band.zlmfit.quality = "Basic"
base_settings.input.band.usesymmetry = "No"
base_settings.input.band.xc.gga = "PBE"
base_settings.input.band.xc.dispersion = "Grimme4"
eda_settings = Settings()
eda_settings.input.band.peda = ""
eda_job = BANDFragmentJob(
fragment1=fromASE(surface),
fragment2=fromASE(adsorbate),
settings=base_settings,
full_settings=eda_settings,
# fragment1_opt=fromASE(surface_opt),
# fragment2_opt=fromASE(adsorbate_opt),
)
eda_job.run()
[28.07|11:20:53] JOB plamsjob STARTED
[28.07|11:20:53] JOB plamsjob RUNNING
[28.07|11:20:53] JOB plamsjob/frag1 STARTED
[28.07|11:20:53] JOB plamsjob/frag1 RUNNING
[28.07|11:21:40] JOB plamsjob/frag1 FINISHED
[28.07|11:21:40] JOB plamsjob/frag1 SUCCESSFUL
[28.07|11:21:40] JOB plamsjob/frag2 STARTED
[28.07|11:21:40] JOB plamsjob/frag2 RUNNING
[28.07|11:21:43] JOB plamsjob/frag2 FINISHED
[28.07|11:21:43] JOB plamsjob/frag2 SUCCESSFUL
... (PLAMS log lines truncated) ...
<scm.plams.recipes.bandfragment.BANDFragmentResults at 0x145404550>
Print Results¶
Finally, we extract the results of the energy decomposition:
results = eda_job.results
eda_res = eda_job.results.get_energy_decomposition(unit="kJ/mol")
print("{:<20} {:>10}".format("Term", "Energy [kJ/mol]"))
for key, value in eda_res.items():
print("{:<20} {:>10.4f}".format(key, value))
print("-------------------------------")
Term Energy [kJ/mol]
E_int -181.9734
E_int_disp -9.1367
E_Pauli 1122.6899
E_elstat -519.8489
E_orb -775.6514
E_1 -1861.0731
E_2 -1.7452
-------------------------------
Complete Python code¶
#!/usr/bin/env amspython
# coding: utf-8
# ## Initialization
from scm.plams import Settings, fromASE, plot_molecule
from scm.plams.recipes.bandfragment import BANDFragmentJob
# build the surface
from ase import Atoms
from ase.build import fcc111, add_adsorbate
# ## Build Surface & Fragments
# We first build a gold surface and add the hydrogen adsorbate.
mol = fcc111("Au", size=(2, 2, 3))
add_adsorbate(mol, "H", 1.5, "ontop")
mol.center(vacuum=10.0, axis=2)
plot_molecule(mol)
# The system is then split into two fragments based on the species.
surface = mol.copy()
symbols = surface.get_chemical_symbols()
del surface[[i for i in range(len(symbols)) if symbols[i] != "Au"]]
adsorbate = mol.copy()
del adsorbate[[i for i in range(len(symbols)) if symbols[i] == "Au"]]
# If available, optimized fragments can also be loaded.
# from ase import io
# surface_opt = io.read("surface_opt.xyz")
# adsorbate_opt = io.read("adsorbate_opt.xyz")
# assert len(surface_opt) == len(surface)
# assert len(adsorbate_opt) == len(adsorbate)
# ## Set Up & Run Job
# For efficiency in this example, we use a minimal basis and reduced computational details to run the job.
base_settings = Settings()
base_settings.input.ams.task = "SinglePoint"
base_settings.input.band.basis.type = "SZ"
base_settings.input.band.basis.core = "Large"
base_settings.input.band.dos.calcdos = "No"
base_settings.input.band.kspace.regular.numberofpoints = "3 3"
base_settings.input.band.beckegrid.quality = "Basic"
base_settings.input.band.zlmfit.quality = "Basic"
base_settings.input.band.usesymmetry = "No"
base_settings.input.band.xc.gga = "PBE"
base_settings.input.band.xc.dispersion = "Grimme4"
eda_settings = Settings()
eda_settings.input.band.peda = ""
eda_job = BANDFragmentJob(
fragment1=fromASE(surface),
fragment2=fromASE(adsorbate),
settings=base_settings,
full_settings=eda_settings,
# fragment1_opt=fromASE(surface_opt),
# fragment2_opt=fromASE(adsorbate_opt),
)
eda_job.run()
# ## Print Results
# Finally, we extract the results of the energy decomposition:
results = eda_job.results
eda_res = eda_job.results.get_energy_decomposition(unit="kJ/mol")
print("{:<20} {:>10}".format("Term", "Energy [kJ/mol]"))
for key, value in eda_res.items():
print("{:<20} {:>10.4f}".format(key, value))
print("-------------------------------")
API¶
- class BANDFragmentJob[source]¶
- Parameters:
- _result_type¶
alias of
BANDFragmentResults
- class BANDFragmentResults[source]¶
Subclass of
ADFFragmentResultsfor BAND calculations.- Parameters:
job (Job) –
- class NOCVBandFragmentJob[source]¶
- _result_type¶
alias of
BANDFragmentResults
- __init__(nocv_settings=None, **kwargs)[source]¶
Create a BANDFragmentJob with final NOCV calculation.
- Parameters:
nocv_settings (Settings, optional) – Settings for the NOCV calculation. Defaults to None.
- Return type:
None
- new_children()[source]¶
After the first round, add the full job to the children list. After the second round, add the NOCV job to the children list.