Kinetic Monte Carlo¶
In this tutorial we will walk you through a Kinetic Monte Carlo (KMC) calculation with the program Zacros, using the GUI. The GUI will generate a pyZacros script that can be run via AMSjobs, just like other runscripts. In addition, with some python experience it should be no problem to extend this script for more advanced tasks.
See also
Zacros documentation in
$AMSHOME/scripting/scm/pyzacros/doc/ZacrosManual.pdf
We will setup the Ziff-Gulari-Barshad model. This will be the same model as in the tutorial on the Zacros website and in the pyZacros tutorial, where more info on the model can be found.
Tip
If you only want to run the simulation without setting it up, download
zacros_tutorial.kin. To open
it, first open AMSkinetics (SCM → Kinetics), select File → Open and browse
to the file.
Step 1: Specify general settings¶
We will start the AMSkinetics module to create our setup. Depending on your license, you might not need to switch to the Zacros program.
In the first step we will specify the general calculation settings. In AMSkinetics they are found on the right panel. These general settings include:
The conditions of the system, such as the temperature and pressure
A random seed, such that multiple runs will produce the same results
The stopping criteria, based on the calculation time, steps, or the real “wall” time (multiple criteria may be specified)
The sampling settings: snapshots, process statistics and species numbers. Thsese can be individually set per event (most useful for test runs) or time step.
All the general settings in the GUI are equivalent to the Zacros input options in simulation_input.dat. These will be generated as pyZacros Settings object in the runscript.
We will use the following settings in our setup:
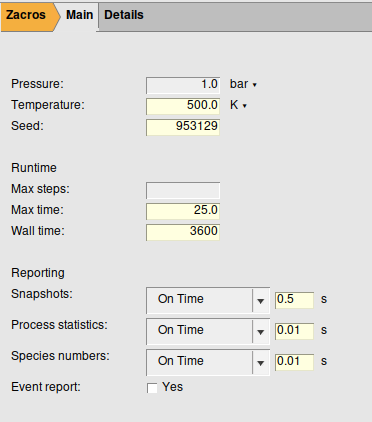
Step 2: Specify the species¶
The gas and surface species can be added on the left panel. In this panel you can switch between the different input settings via the tabs. In this step we will start with the Gas species tab.
With the  you can add new gas species. You can populate fields with values by hand, or paste preformatted data from a spreadsheet.
If fields are left empty, a default value of 0.0 eV will be used for the gas energy and 0.0 for the initial molar fraction.
you can add new gas species. You can populate fields with values by hand, or paste preformatted data from a spreadsheet.
If fields are left empty, a default value of 0.0 eV will be used for the gas energy and 0.0 for the initial molar fraction.
We will use the following gas species in our setup:
Next, we will add the surface species on the ‘Surface species’ tab. By default, there is always the empty site *. The surface species names conventionally have * characters at the end to denote the denticity of the species. AMSkinetics will fill in the denticity correctly if the surface species names follow this convention.
In later steps, the gas and surface species will be used to set up the clusters and reactions, via their respective tabs.
Note that it is still possible to modify or add new species after setting these up.
In Zacros, the species are part of the simulation_input.dat file and are handled in the same way.
In pyZacros, they will be generated as pyZacros Species in the runscript, whilst the molar fractions are part of the Settings object, to aid more advanced scripting.
Step 3: Specify the lattice structure¶
The lattice structure is found on the middle panel. Here you will find tools to build the lattice. To create a new lattice, simply select a Preset lattice from the list, specify the Lattice constant and click Build (note, however, that this will delete any created clusters and reactions, as these may depend on the deleted sites).
The preset lattices can also be further modified and will be passed on to Zacros as a “Unit-Cell-Defined Periodic Lattice”. For example, the lattice Vectors can have custom values and the Repeats can also be modified at any point. A repeat of 1 can be used if a complete custom lattice is desired.
Finally it is possible to create new Site types and add new Sites.
On the right is an interactive lattice display. The following actions can be used to navigate this display:
Below the lattice display, there are also buttons to add or remove connections between selected sites.
We will build the lattice using these steps:
In Zacros the lattice settings are in the lattice_input.dat. They will be generated as a pyZacros Lattice in the runscript.
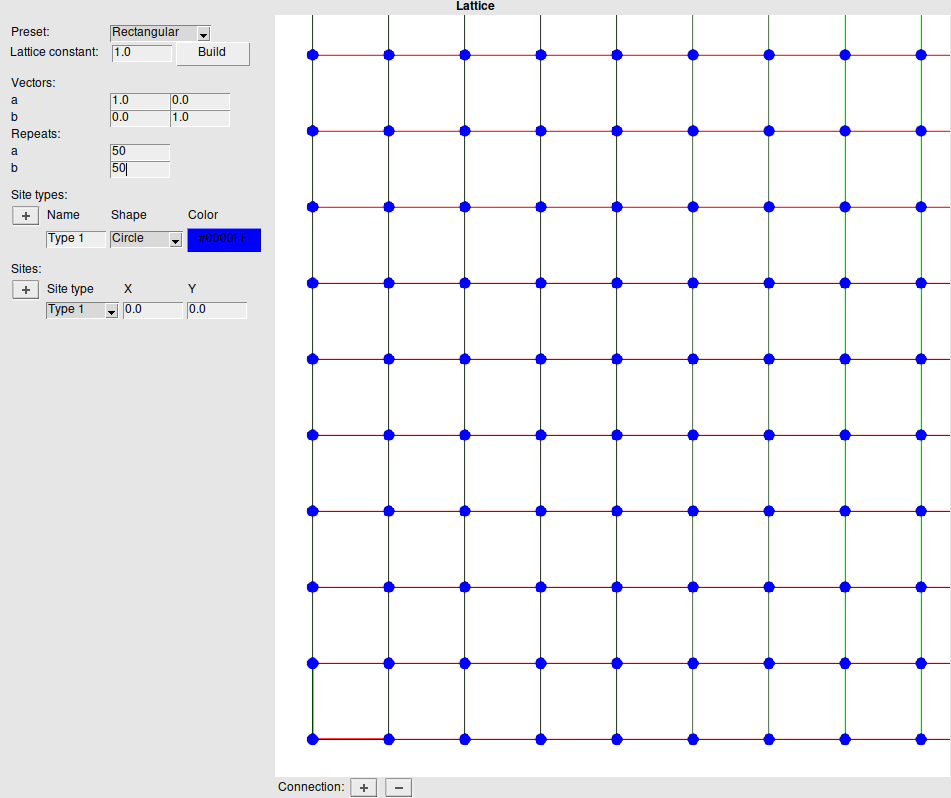
Step 4: Specify the energetics model: Clusters¶
Clusters are found on the left panel under the Clusters tab. Clusters are represented as a graph. We can define these by selecting one or more connected sites from the lattice. This makes sure the graph actually exists in the defined lattice, and immediately gives a good visualization of the graph. After selecting the pattern you can click the  to add a new cluster.
to add a new cluster.
The Multiplicity counts the amount of times the representation is found on the lattice. It will therefore divide the Cluster energy, but can be kept at 1 if the cluster energy is already normalized.
Finally the Species can be assigned to the selected sites. Multidentate species should have the same Entity number on each bound site.
When saving the final setup, the GUI will check if this is consistent. You can Show the cluster on the lattice, which selects and labels the sites. The sites closest to the origin are used to show the cluster representation. You can also Update the cluster to change the graph to the new selection, or Delete to remove the cluster.
We will add two clusters, for CO* and O*:
 .
.
In Zacros the cluster settings are in the energetics_input.dat file. They will be generated as pyZacros Clusters.
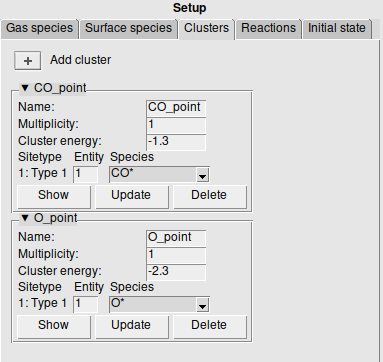
Step 5: Specify the reaction mechanism: Reactions¶
Reactions are found on the left panel under the Reactions tab. Just like clusters, reactions are represented as a graph, and are defined via the same method. Each site now represents the initial and final state of an elementary event.
When adding a reaction, the Reversible checkbox indicates whether the reaction is reversible. The Pre exponential specifies the pre-exponential for the forward reaction in the Arrhenius formula. For reversible reactions the PE ratio specifies the ratio between the forward and reverse pre-exponentials. The Activation energy specifies the activation energy at the zero coverage limit for the forward reaction.
For each site the Initial and Final species can be specified. Again, like for the clusters, the Entity number should be the same for multidentate species. Additional Gas species can be specified for each side of the reaction. The Show, Update, and Delete work the same as for the clusters.
We will add three reactions; two adsorption reactions and the oxidation reaction:
 .
. .
. .
.In Zacros the reaction mechanism settings are in the mechanism_input.dat file. They will be generated as a pyZacros ElementaryReaction.
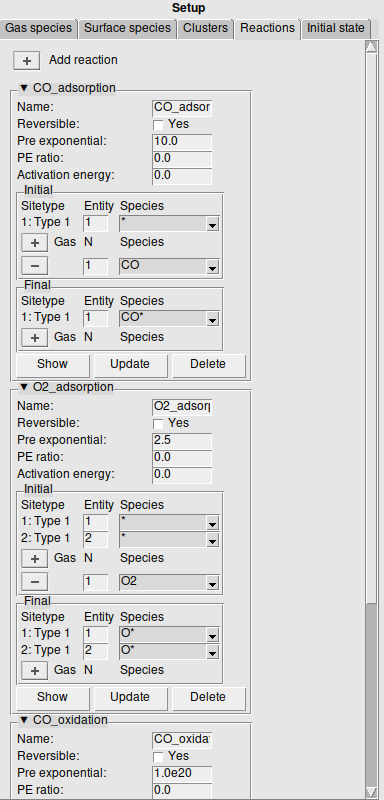
Step 6: Run and analyze the results¶
The Initial state tab on the left panel is not used in this tutorial. It can be used to either randomly or specifically populate sites with surface species for the initial state.
We will now save and run the setup. This will generate a file ending with .kin which is used to store the GUI setup, which can be opened again later by AMSkinetics. This is analogous to the .ams files for AMSinput. The runscript is as usual saved in a .run file, and contains the full pyZacros script.
All the files that pyZacros generates and the output of Zacros will end up inside the .results directory. If the AMSkinetics module is still open, it will now ask to analyze the results. You can also analyze the results via View → Results.
This will bring up Zacros-post, a program for analyzing Zacros results. If you do not have Zacros-post installed, it will prompt you to install it. Zacros-post can also be installed or uninstalled using the SCM package manager (SCM → Packages).
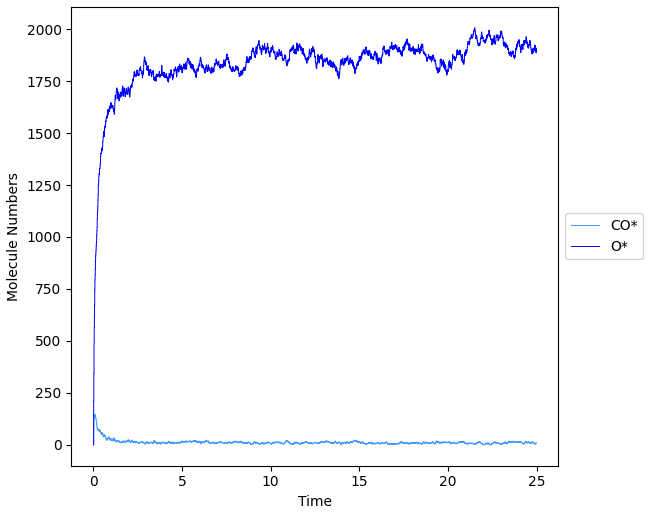
In Zacros-post, you can for example select Plot → Species Numbers to see the surface species as a function of time
Other examples¶
Download
zacros_reversible.kinfor an example with reversible reactions, based on A Simple KMC simulation from zacros.org