Molecule substitution: Attach ligands to substrates¶
Script showing how to created substituted benzene molecules using PLAMS, by combining a benzene molecule with a different molecule and defining one bond on each molecule (the “connector”) to break and where a new bond will be formed.
See also
Note: This example requires AMS2023 or later.
To follow along, either
Download
MoleculeSubstitution.py(run as$AMSBIN/amspython MoleculeSubstitution.py).Download
MoleculeSubstitution.ipynb(see also: how to install Jupyterlab in AMS)
Worked Example¶
Initial imports¶
import scm.plams as plams
import matplotlib.pyplot as plt
from rdkit import Chem
from rdkit.Chem import Draw
from rdkit.Chem.Draw import IPythonConsole
from rdkit.Chem import AllChem
from typing import List, Tuple
IPythonConsole.ipython_useSVG = True
IPythonConsole.molSize = 250, 250
# this line is not required in AMS2025+
plams.init()
PLAMS working folder: /path/plams/examples/MoleculeSubstitution/plams_workdir
Helper class and function¶
The MoleculeConnector class and substitute() method below are convenient to use.
class MoleculeConnector:
def __init__(self, molecule, connector, name="molecule"):
self.name = name
self.molecule = molecule
self.molecule.properties.name = name
self.connector = connector # 2-tuple of integers, unlike the Molecule.substitute() method which uses two atoms
def __str__(self):
return f"""
Name: {self.name}
{self.molecule}
Connector: {self.connector}. This means that when substitution is performed atom {self.connector[0]} will be kept in the substituted molecule. Atom {self.connector[1]}, and anything connected to it, will NOT be kept.
"""
def substitute(substrate: MoleculeConnector, ligand: MoleculeConnector):
"""
Returns: Molecule with the ligand added to the substrate, replacing the respective connector bonds.
"""
molecule = substrate.molecule.copy()
molecule.substitute(
connector=(molecule[substrate.connector[0]], molecule[substrate.connector[1]]),
ligand=ligand.molecule,
ligand_connector=(ligand.molecule[ligand.connector[0]], ligand.molecule[ligand.connector[1]]),
)
return molecule
def set_atom_indices(rdmol: Chem.rdchem.Mol, start=0):
for atom in rdmol.GetAtoms():
atom.SetAtomMapNum(atom.GetIdx() + start) # give 1-based index
return rdmol
def to_lewis(molecule: plams.Molecule, template=None, regenerate: bool = True):
if isinstance(molecule, Chem.rdchem.Mol):
rdmol = molecule
else:
rdmol = plams.to_rdmol(molecule)
if regenerate:
rdmol = Chem.RemoveHs(rdmol)
smiles = Chem.MolToSmiles(rdmol)
rdmol = Chem.MolFromSmiles(smiles)
if template is not None:
AllChem.GenerateDepictionMatching2DStructure(rdmol, template)
try:
if molecule.properties.name:
rdmol.SetProp("_Name", molecule.properties.name)
except AttributeError:
pass
return rdmol
def smiles2template(smiles: str):
template = Chem.MolFromSmiles(smiles)
AllChem.Compute2DCoords(template)
return template
def draw_lewis_grid(
molecules: List[plams.Molecule],
molsPerRow: int = 4,
template_smiles: str = None,
regenerate: bool = False,
draw_atom_indices: bool = False,
draw_legend: bool = True,
):
template = None
if template_smiles:
template = smiles2template(template_smiles)
rdmols = [to_lewis(x, template=template, regenerate=regenerate) for x in molecules]
if draw_atom_indices:
for rdmol in rdmols:
set_atom_indices(rdmol, start=1)
legends = None
if draw_legend:
try:
legends = [x.properties.name or f"mol{i}" for i, x in enumerate(molecules)]
except AttributeError:
pass
return Draw.MolsToGridImage(rdmols, molsPerRow=molsPerRow, legends=legends)
def view_molecules(molecules: List[plams.Molecule], titles: List[str], figsize: Tuple[int], **kwargs):
fig, axes = plt.subplots(1, len(molecules), figsize=figsize)
if len(molecules) == 1:
axes = [axes]
try:
from scm.plams import view # view molecule using AMSview in a Jupyter Notebook in AMS2026+
imgs = [view(m, **kwargs) for m in molecules]
for ax, img, title in zip(axes, imgs, titles):
ax.imshow(img)
ax.axis("off")
ax.set_title(title)
plt.show()
except ImportError:
from scm.plams import plot_molecule # plot molecule in a Jupyter Notebook in AMS2023+
for ax, mol, title in zip(axes, molecules, titles):
plot_molecule(mol, ax=ax)
ax.set_title(title)
def view_molecule(molecule: plams.Molecule, title: str, figsize: Tuple[int] = (8, 8), **kwargs):
view_molecules([molecule], [title], figsize, **kwargs)
Generate substrate molecule¶
substrate_smiles = "c1ccccc1"
substrate = plams.from_smiles(substrate_smiles, forcefield="uff")
substrate.properties.name = "benzene"
Find out which bond to cleave¶
In the molecule you need to define which bond to cleave. To find out the bonds, run for example:
for b in substrate.bonds:
el1 = b.atom1.symbol
el2 = b.atom2.symbol
idx1, idx2 = substrate.index(b)
print(f"{el1}({idx1})--{el2}({idx2})")
C(1)--C(2)
C(2)--C(3)
C(3)--C(4)
C(4)--C(5)
C(5)--C(6)
C(6)--C(1)
C(1)--H(7)
C(2)--H(8)
C(3)--H(9)
C(4)--H(10)
C(5)--H(11)
C(6)--H(12)
to find that atoms 6 (C) and 12 (H) are bonded. We will choose this bond to cleave.
Alternatively, we can inspect the molecule inside a Jupyter notebook, visualizing with AMSView, to also find that atoms 6 (C) and 12 (H) are bonded.
view_molecule(substrate, substrate.properties.name, padding=-0.5, show_atom_labels=True, atom_label_type="Name")
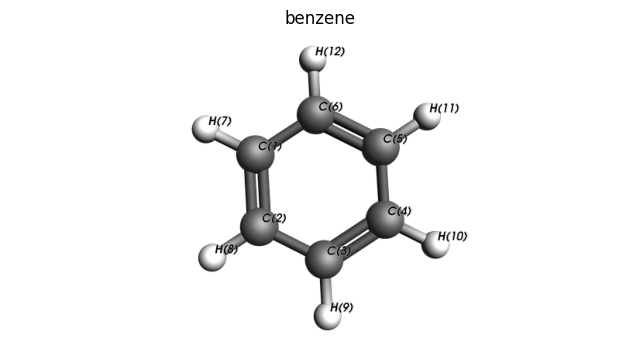
substrate_connector = MoleculeConnector(
substrate, (6, 12), "phenyl"
) # benzene becomes phenyl when bond between atoms 6,12 is cleaved
Define ligands¶
Perform the same steps for the ligands.
Note: The ligands below have an extra hydrogen or even more atoms compared to the name that they’re given.
ligands = [
MoleculeConnector(
plams.from_smiles("CCOC(=O)C", forcefield="uff"), (3, 2), "acetate"
), # ethyl acetate, bond from O to C cleaved
MoleculeConnector(
plams.from_smiles("O=NO", forcefield="uff"), (3, 4), "nitrite"
), # nitrous acid, bond from O to H cleaved
MoleculeConnector(
plams.from_smiles("Cl", forcefield="uff"), (1, 2), "chloride"
), # hydrogen chloride, bond from Cl to H cleaved
MoleculeConnector(
plams.from_smiles("c1ccccc1", forcefield="uff"), (6, 12), "phenyl"
), # benzene, bond to C to H cleaved
]
ligand_molecules = [ligand.molecule for ligand in ligands]
view_molecules(
ligand_molecules,
[ligand.name for ligand in ligands],
figsize=(15, 4),
width=400,
show_atom_labels=True,
atom_label_type="Name",
)
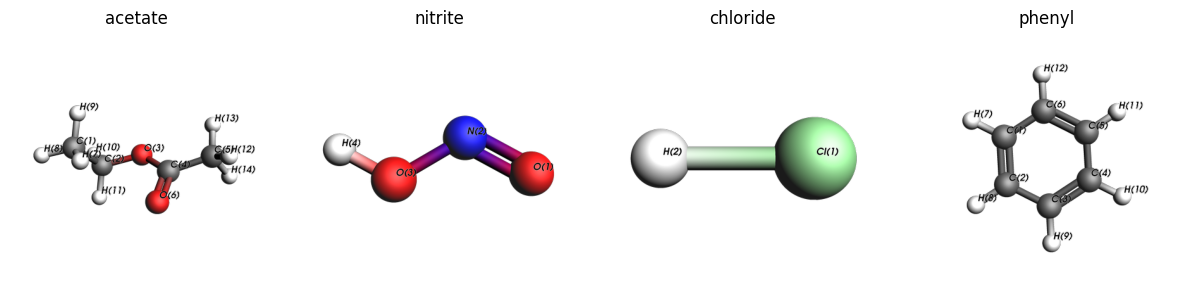
Above we see that cleaving the bonds from O(3)-C(2), O(3)-H(4), Cl(1)-H(2), and C(6)-H(12) will give the acetate, nitrite, chloride, and phenyl substituents, respectively.
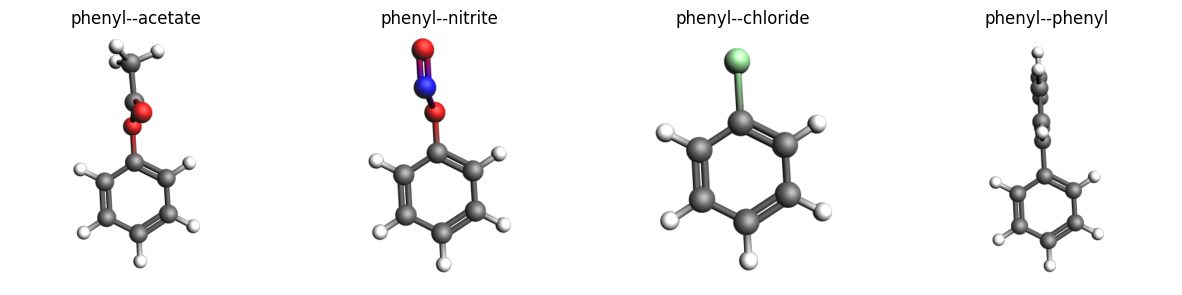
Generate substituted molecules¶
mols = []
for ligand in ligands:
mol = substitute(substrate_connector, ligand)
mol.properties.name = f"{substrate_connector.name}--{ligand.name}"
mols.append(mol)
print(f"Writing {mol.properties.name}.xyz")
mol.write(f"{mol.properties.name}.xyz")
print(f"{mol.properties.name} formula: {mol.get_formula(as_dict=True)}")
Writing phenyl--acetate.xyz
phenyl--acetate formula: {'C': 8, 'H': 8, 'O': 2}
Writing phenyl--nitrite.xyz
phenyl--nitrite formula: {'C': 6, 'H': 5, 'O': 2, 'N': 1}
Writing phenyl--chloride.xyz
phenyl--chloride formula: {'C': 6, 'H': 5, 'Cl': 1}
Writing phenyl--phenyl.xyz
phenyl--phenyl formula: {'C': 12, 'H': 10}
Plot 3D structures with PLAMS¶
view_molecules(mols, [mol.properties.name for mol in mols], figsize=(15, 4), width=400, padding=-0.3)

Plot 2D Lewis structures with RDKit¶
The molecules can be aligned by using a benzene template. The regenerate option regenerates the molecule with RDkit to clean up the atomic positions.
draw_lewis_grid(mols, template_smiles=substrate_smiles, regenerate=True)
Complete Python code¶
#!/usr/bin/env amspython
# coding: utf-8
# ## Initial imports
import scm.plams as plams
import matplotlib.pyplot as plt
from rdkit import Chem
from rdkit.Chem import Draw
from rdkit.Chem.Draw import IPythonConsole
from rdkit.Chem import AllChem
from typing import List, Tuple
IPythonConsole.ipython_useSVG = True
IPythonConsole.molSize = 250, 250
# this line is not required in AMS2025+
plams.init()
# ## Helper class and function
#
# The ``MoleculeConnector`` class and ``substitute()`` method below are convenient to use.
class MoleculeConnector:
def __init__(self, molecule, connector, name="molecule"):
self.name = name
self.molecule = molecule
self.molecule.properties.name = name
self.connector = connector # 2-tuple of integers, unlike the Molecule.substitute() method which uses two atoms
def __str__(self):
return f"""
Name: {self.name}
{self.molecule}
Connector: {self.connector}. This means that when substitution is performed atom {self.connector[0]} will be kept in the substituted molecule. Atom {self.connector[1]}, and anything connected to it, will NOT be kept.
"""
def substitute(substrate: MoleculeConnector, ligand: MoleculeConnector):
"""
Returns: Molecule with the ligand added to the substrate, replacing the respective connector bonds.
"""
molecule = substrate.molecule.copy()
molecule.substitute(
connector=(molecule[substrate.connector[0]], molecule[substrate.connector[1]]),
ligand=ligand.molecule,
ligand_connector=(ligand.molecule[ligand.connector[0]], ligand.molecule[ligand.connector[1]]),
)
return molecule
def set_atom_indices(rdmol: Chem.rdchem.Mol, start=0):
for atom in rdmol.GetAtoms():
atom.SetAtomMapNum(atom.GetIdx() + start) # give 1-based index
return rdmol
def to_lewis(molecule: plams.Molecule, template=None, regenerate: bool = True):
if isinstance(molecule, Chem.rdchem.Mol):
rdmol = molecule
else:
rdmol = plams.to_rdmol(molecule)
if regenerate:
rdmol = Chem.RemoveHs(rdmol)
smiles = Chem.MolToSmiles(rdmol)
rdmol = Chem.MolFromSmiles(smiles)
if template is not None:
AllChem.GenerateDepictionMatching2DStructure(rdmol, template)
try:
if molecule.properties.name:
rdmol.SetProp("_Name", molecule.properties.name)
except AttributeError:
pass
return rdmol
def smiles2template(smiles: str):
template = Chem.MolFromSmiles(smiles)
AllChem.Compute2DCoords(template)
return template
def draw_lewis_grid(
molecules: List[plams.Molecule],
molsPerRow: int = 4,
template_smiles: str = None,
regenerate: bool = False,
draw_atom_indices: bool = False,
draw_legend: bool = True,
):
template = None
if template_smiles:
template = smiles2template(template_smiles)
rdmols = [to_lewis(x, template=template, regenerate=regenerate) for x in molecules]
if draw_atom_indices:
for rdmol in rdmols:
set_atom_indices(rdmol, start=1)
legends = None
if draw_legend:
try:
legends = [x.properties.name or f"mol{i}" for i, x in enumerate(molecules)]
except AttributeError:
pass
return Draw.MolsToGridImage(rdmols, molsPerRow=molsPerRow, legends=legends)
def view_molecules(molecules: List[plams.Molecule], titles: List[str], figsize: Tuple[int], **kwargs):
fig, axes = plt.subplots(1, len(molecules), figsize=figsize)
if len(molecules) == 1:
axes = [axes]
try:
from scm.plams import view # view molecule using AMSview in a Jupyter Notebook in AMS2026+
imgs = [view(m, **kwargs) for m in molecules]
for ax, img, title in zip(axes, imgs, titles):
ax.imshow(img)
ax.axis("off")
ax.set_title(title)
plt.show()
except ImportError:
from scm.plams import plot_molecule # plot molecule in a Jupyter Notebook in AMS2023+
for ax, mol, title in zip(axes, molecules, titles):
plot_molecule(mol, ax=ax)
ax.set_title(title)
def view_molecule(molecule: plams.Molecule, title: str, figsize: Tuple[int] = (8, 8), **kwargs):
view_molecules([molecule], [title], figsize, **kwargs)
# ## Generate substrate molecule
substrate_smiles = "c1ccccc1"
substrate = plams.from_smiles(substrate_smiles, forcefield="uff")
substrate.properties.name = "benzene"
# ## Find out which bond to cleave
# In the molecule you need to define which bond to cleave. To find out the bonds, run for example:
for b in substrate.bonds:
el1 = b.atom1.symbol
el2 = b.atom2.symbol
idx1, idx2 = substrate.index(b)
print(f"{el1}({idx1})--{el2}({idx2})")
# to find that atoms 6 (C) and 12 (H) are bonded. We will choose this bond to cleave.
#
# Alternatively, we can inspect the molecule inside a Jupyter notebook, visualizing with AMSView, to also find that atoms 6 (C) and 12 (H) are bonded.
view_molecule(substrate, substrate.properties.name, padding=-0.5, show_atom_labels=True, atom_label_type="Name")
substrate_connector = MoleculeConnector(
substrate, (6, 12), "phenyl"
) # benzene becomes phenyl when bond between atoms 6,12 is cleaved
# ## Define ligands
# Perform the same steps for the ligands.
#
# **Note**: The ligands below have an extra hydrogen or even more atoms compared to the name that they're given.
ligands = [
MoleculeConnector(
plams.from_smiles("CCOC(=O)C", forcefield="uff"), (3, 2), "acetate"
), # ethyl acetate, bond from O to C cleaved
MoleculeConnector(
plams.from_smiles("O=NO", forcefield="uff"), (3, 4), "nitrite"
), # nitrous acid, bond from O to H cleaved
MoleculeConnector(
plams.from_smiles("Cl", forcefield="uff"), (1, 2), "chloride"
), # hydrogen chloride, bond from Cl to H cleaved
MoleculeConnector(
plams.from_smiles("c1ccccc1", forcefield="uff"), (6, 12), "phenyl"
), # benzene, bond to C to H cleaved
]
ligand_molecules = [ligand.molecule for ligand in ligands]
view_molecules(
ligand_molecules,
[ligand.name for ligand in ligands],
figsize=(15, 4),
width=400,
show_atom_labels=True,
atom_label_type="Name",
)
# Above we see that cleaving the bonds from O(3)-C(2), O(3)-H(4), Cl(1)-H(2), and C(6)-H(12) will give the acetate, nitrite, chloride, and phenyl substituents, respectively.
# ## Generate substituted molecules
mols = []
for ligand in ligands:
mol = substitute(substrate_connector, ligand)
mol.properties.name = f"{substrate_connector.name}--{ligand.name}"
mols.append(mol)
print(f"Writing {mol.properties.name}.xyz")
mol.write(f"{mol.properties.name}.xyz")
print(f"{mol.properties.name} formula: {mol.get_formula(as_dict=True)}")
# ## Plot 3D structures with PLAMS
view_molecules(mols, [mol.properties.name for mol in mols], figsize=(15, 4), width=400, padding=-0.3)
# ## Plot 2D Lewis structures with RDKit
#
# The molecules can be aligned by using a benzene template. The ``regenerate`` option regenerates the molecule with RDkit to clean up the atomic positions.
draw_lewis_grid(mols, template_smiles=substrate_smiles, regenerate=True)