PLAMS
Python scripting to streamline your workflows
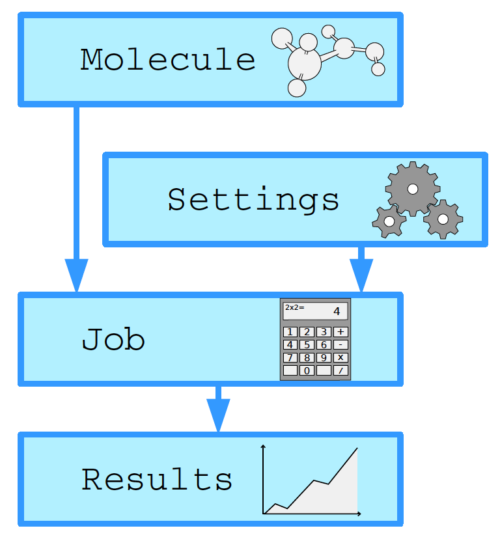
The Python Library for Automating Molecular Simulation (PLAMS) is powerful and flexible Python tool interfaced to the Amsterdam Modeling Suite engines ADF, BAND, DFTB, MOPAC, ReaxFF, and UFF. PLAMS is easily extendable and interfaced to external molecular modeling programs.
Getting started with PLAMS
SCM’s expert Mirko Franchini demonstrates how the PLAMS library can be used to automatize workflows and do high-throughput calculations.