DNA/RNA transverse current sequencing: Intrinsic structural noise from neighboring bases
Nanopore DNA sequencing via transverse current offers a number of advantages with respect to current technologies, however, the base calling error rates are still too high. The source of noise has to be understood in order to devise methods to decrease them. In a recent paper an effective multi-orbital tight-binding model derived from first-principles ADF calculations has been used to calculate the transverse current through DNA/RNA molecules. A very important source of error in identifying nucleotides come from current through neighboring bases due to carrier dispersion along the chain of the molecule, causing the smearing of the current distributions. At the same time this implies that currents through neighboring bases are correlated. These correlations can be used to account for satelite contributions and thereby make the current distributions more distinguishable, thus reducing the error rates.
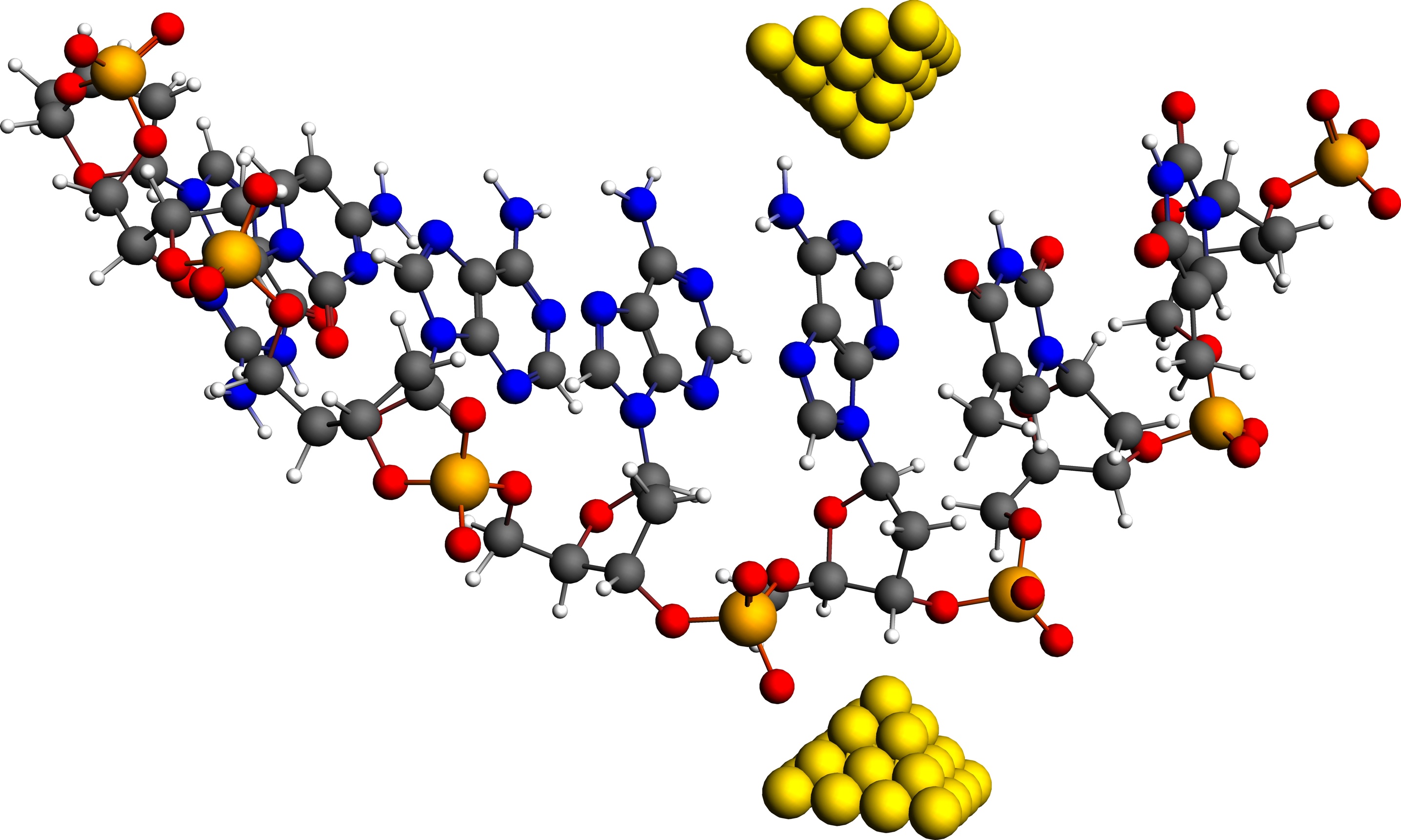
Schematic representation of transverse current DNA sequencing setup: A single-strand DNA molecule translocating between metal contacts embedded in a nanopore.
Fragment Analysis, Fragment Molecular Orbitals, Charge Transport
J. Alvarez, D. Skachkov, S. Massey, A. Kalitsov, and J. Velev, DNA/RNA transverse current sequencing: Intrinsic structural noise from neighboring bases, Frontiers in Genetics 6, 213 (2015)
Key conceptsADF bonding analysis Charge Transport pharma