Modeling the hydrogen bonds, pi-stacking and solvent effects in DNA
In a recent paper in ChemComm, researchers in Spain and the Netherlands have used a combination of ADF features to unravel the factors behind the fidelity in chemical, template-assisted primer extension. The analyses therein, based on state-of-the-art dispersion-corrected density functional theory (DFT), reveal key factors behind the intrinsic affinity of a DNA template-primer complex to select the correct nucleotide. They show that hydrogen bonds alone are not enough to achieve the correct preference for Watson-Crick pairs. This correct preference is only achieved after including solvent effects (partial desolvation). Stacking interactions, entropy effects and the helical structure imposed by the backbone are also crucial. Interestingly, the researchers also find what they dubbed the “nearest-neighbor effect”: the nature of the terminal base in the primer strand has a non-negligible effect on the energetic preference of the template-primer complex for the correct incoming nucleotide. In other words, the precision of a particular DNA replication step fluctuates as a function of which nucleotide was incorporated in the previous cycle.
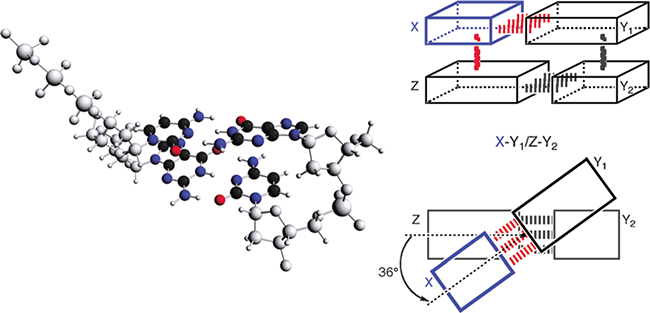
Above: A model complex of incoming nucleotide X’ + template Y1‘/Y2‘ strand + primer strand Z’ in aqueous solution (see the text of the publication for details).
The features of ADF which made this study possible include the ability to treat simultaneously covalent and dispersion interactions (using Grimme’s dispersion corrections) along with treatment of solvent effects (with COSMO).
J. Poater, M. Swart, C.Fonseca Guerra and F. M. Bickelhaupt, Selectivity in DNA replication. Interplay of steric shape, hydrogen bonds, n-stacking and solvent effects. Chemical Communications 47, 7326 (2011).
Key conceptsADF bonding analysis Dispersion pharma solvation