Ziff-Gulari-Barshad model: Phase Transitions under Steady State Conditions.¶
Note
To follow this tutorial, either:
Download
PhaseTransitions-SteadyState.py(run as$AMSBIN/amspython PhaseTransitions-SteadyState.py).Download
PhaseTransitions-SteadyState.ipynb(see also: how to install Jupyterlab)
This example is inspired in the seminal paper: Kinetic Phase Transitions in an Irreversible Surface-Reaction Model by Robert M. Ziff, Erdagon Gulari, and Yoav Barshad in 1986 Phys. Rev. Lett. 56, (1986) 2553. The authors proposed a simple model for catalytic reactions of carbon monoxide oxidation to carbon dioxide on a surface. This model is now known as the Ziff-Gulari-Barshad (ZGB) model after their names. While the model leaves out many important steps of the real system, it exhibits interesting steady-state off-equilibrium behavior and two types of phase transitions, which actually occur in real systems. Please refer to the original paper for more details. In this example, we will analyze the effect of changing the composition of the gas phase, namely partial pressures for \(O_2\) and \(CO\), in the \(CO_2\) Turnover frequency (TOF) in the ZGB model. At variance with the example Phase Transitions in the ZGB model, here we extend the dynamics up to reaching the system’s steady state for each gas phase composition value. Most of the code is the same, except for the section regarding the setup of the steady state calculation.
First, we import all packages we need:
import multiprocessing
import numpy
import scm.plams
import scm.pyzacros as pz
import scm.pyzacros.models
Then, we initialize the pyZacros environment:
scm.pyzacros.init()
PLAMS working folder: /home/user/pyzacros/examples/ZiffGulariBarshad/plams_workdir
On a typical laptop, this calculation should take no more than 10 min to
complete. Here we illustrate how to run several parallel Zacros
calculations. We’ll use the plams.JobRunner class, which easily
allows us to run as many parallel instances as we request. In this case,
we choose to use the maximum number of simultaneous processes
(maxjobs) equal to the number of processors in the machine.
Additionally, by setting nproc = 1 we establish that only one
processor will be used for each zacros instance.
maxjobs = multiprocessing.cpu_count()
scm.plams.config.default_jobrunner = scm.plams.JobRunner(parallel=True, maxjobs=maxjobs)
scm.plams.config.job.runscript.nproc = 1
print('Running up to {} jobs in parallel simultaneously'.format(maxjobs))
Running up to 8 jobs in parallel simultaneously
Now, we initialize our Ziff-Gulari-Barshad model, which by luck is available as a predefined model in pyZacros,
zgb = pz.models.ZiffGulariBarshad()
Then, we must set up a ZacrosParametersScanJob calculation, which
will allow us to scan the molar fraction of \(CO\) as a parameter.
However, this calculation requires the definition of a
ZacrosSteadyStateJob, that in turns requires a ZacrosJob. So, We
will go through them one at a time:
1. Setting up the ZacrosJob
For ZacrosJob, all parameters are set using a Setting object. To
begin, we define the physical parameters: temperature (in K), and
pressure (in bar). The calculation parameters are then set:
species numbers (in s) determines how frequently information about
the number of gas and surface species will be stored, max time (in
s) specifies the maximum allowed simulated time, and “random seed”
specifies the random seed to make the calculation precisely
reproducible. Keep in mind that max time defines the calculation’s
stopping criterion, and it is the parameter that will be controlled
later to achieve the steady-state configuration. Finally, we create the
ZacrosJob, which uses the parameters we just defined as well as the
Ziff-Gulari-Barshad model’s lattice, mechanism, and cluster expansion.
Notice we do not run this job, we use it as a reference for the
steady-state calculation described below.
z_sett = pz.Settings()
z_sett.temperature = 500.0
z_sett.pressure = 1.0
z_sett.max_time = 10.0
z_sett.species_numbers = ('time', 0.1)
z_sett.random_seed = 953129
z_job = pz.ZacrosJob( settings=z_sett, lattice=zgb.lattice,
mechanism=zgb.mechanism,
cluster_expansion=zgb.cluster_expansion )
2. Setting up the ZacrosSteadyStateJob
We also need to create a Setting object for ZacrosJob There, we
ask for a steady-state configuration using a TOFs calculation with a 96%
confidence level (turnover frequency.confidence), using four
replicas to speed up the calculation (turnover frequency.nreplicas);
for more information, see example Ziff-Gulari-Barshad model: Steady
State Conditions. The ZacrosSteadyStateJob.Parameters object
allows to set the grid of maximum times to explore in order to reach the
steady state (ss_params). Finally, we create
ZacrosSteadyStateJob, which references the ZacrosJob defined
above (z_job) as well as the Settings object and parameters we
just defined:
ss_sett = pz.Settings()
ss_sett.turnover_frequency.nbatch = 20
ss_sett.turnover_frequency.confidence = 0.96
ss_sett.turnover_frequency.nreplicas = 4
ss_params = pz.ZacrosSteadyStateJob.Parameters()
ss_params.add( 'max_time', 'restart.max_time',
2*z_sett.max_time*( numpy.arange(10)+1 )**2 )
ss_job = pz.ZacrosSteadyStateJob( settings=ss_sett, reference=z_job,
parameters=ss_params )
[02.02|22:26:23] JOB plamsjob Steady State Convergence: Using nbatch=20,confidence=0.96,ignore_nbatch=1,nreplicas=4
3. Setting up the ZacrosParametersScanJob
Although the ZacrosParametersScanJob does not require a Setting
object, it does require a ZacrosSteadyStateJob.Parameters object to
specify which parameters must be modified systematically. In this
instance, all we need is a dependent parameter, the \(O_2\) molar
fraction x_O2, and an independent parameter, the \(CO\) molar
fraction x_CO, which ranges from 0.2 to 0.8 in steps of 0.01. Keep
in mind that the condition x_CO+x_O2=1 must be met. These molar
fractions will be used internally to replace molar fraction.CO and
molar fraction.O2 in the Zacros input files. Then, using the
ZacrosSteadyStateJob defined earlier (ss job) and the parameters
we just defined (ps params), we create the
ZacrosParametersScanJob:
ps_params = pz.ZacrosParametersScanJob.Parameters()
ps_params.add( 'x_CO', 'molar_fraction.CO', numpy.arange(0.2, 0.8, 0.01) )
ps_params.add( 'x_O2', 'molar_fraction.O2', lambda params: 1.0-params['x_CO'] )
ps_job = pz.ZacrosParametersScanJob( reference=ss_job, parameters=ps_params )
[02.02|22:26:23] JOB ps_cond000 Steady State Convergence: Using nbatch=20,confidence=0.96,ignore_nbatch=1,nreplicas=4
[02.02|22:26:23] JOB ps_cond001 Steady State Convergence: Using nbatch=20,confidence=0.96,ignore_nbatch=1,nreplicas=4
[02.02|22:26:23] JOB ps_cond002 Steady State Convergence: Using nbatch=20,confidence=0.96,ignore_nbatch=1,nreplicas=4
[02.02|22:26:23] JOB ps_cond003 Steady State Convergence: Using nbatch=20,confidence=0.96,ignore_nbatch=1,nreplicas=4
[02.02|22:26:23] JOB ps_cond004 Steady State Convergence: Using nbatch=20,confidence=0.96,ignore_nbatch=1,nreplicas=4
...
[02.02|22:26:23] JOB ps_cond056 Steady State Convergence: Using nbatch=20,confidence=0.96,ignore_nbatch=1,nreplicas=4
[02.02|22:26:23] JOB ps_cond057 Steady State Convergence: Using nbatch=20,confidence=0.96,ignore_nbatch=1,nreplicas=4
[02.02|22:26:23] JOB ps_cond058 Steady State Convergence: Using nbatch=20,confidence=0.96,ignore_nbatch=1,nreplicas=4
[02.02|22:26:23] JOB ps_cond059 Steady State Convergence: Using nbatch=20,confidence=0.96,ignore_nbatch=1,nreplicas=4
[02.02|22:26:23] JOB ps_cond060 Steady State Convergence: Using nbatch=20,confidence=0.96,ignore_nbatch=1,nreplicas=4
The parameters scan calculation setup is ready. Therefore, we can start
it by invoking the function run(), which will provide access to the
results via the results variable after it has been completed. The
sentence involving the method ok(), verifies that the calculation
was successfully executed, and waits for the completion of every
executed thread. This step should take less than 10 mins!
results = ps_job.run()
if not results.job.ok():
print('Something went wrong!')
[02.02|22:26:23] JOB plamsjob STARTED
[02.02|22:26:23] Waiting for job plamsjob to finish
[02.02|22:26:23] JOB plamsjob RUNNING
[02.02|22:26:23] JOB plamsjob/ps_cond000 STARTED
[02.02|22:26:23] JOB plamsjob/ps_cond001 STARTED
[02.02|22:26:23] JOB plamsjob/ps_cond002 STARTED
...
[02.02|22:29:57] CO2 0.44695 0.02674 0.05983 False
[02.02|22:29:57] JOB plamsjob/ps_cond020/ss_iter004_rep000 FINISHED
[02.02|22:29:57] JOB plamsjob/ps_cond020/ss_iter004_rep000 FAILED
[02.02|22:29:57] Waiting for job ss_iter004_rep001 to finish
[02.02|22:29:57] JOB plamsjob/ps_cond020/ss_iter004_rep001 FINISHED
[02.02|22:29:58] JOB plamsjob/ps_cond020/ss_iter004_rep002 FINISHED
[02.02|22:29:58] Average
[02.02|22:29:58] JOB plamsjob/ps_cond020/ss_iter004_rep001 SUCCESSFUL
[02.02|22:29:58] Waiting for job ss_iter004_rep003 to finish
[02.02|22:29:58] species TOF error ratio conv?
[02.02|22:29:58] CO -0.42282 0.01585 0.03748 True
[02.02|22:29:58] O2 -0.21130 0.00793 0.03751 True
[02.02|22:29:58] CO2 0.42282 0.01585 0.03748 True
[02.02|22:29:58] JOB plamsjob/ps_cond021 Steady State Convergence: CONVERGENCE REACHED. DONE!
[02.02|22:29:58] JOB plamsjob/ps_cond020/ss_iter004_rep002 SUCCESSFUL
[02.02|22:29:58] JOB plamsjob/ps_cond020/ss_iter004_rep003 FINISHED
[02.02|22:29:58] JOB plamsjob/ps_cond021 FINISHED
[02.02|22:29:59] JOB plamsjob/ps_cond020/ss_iter004_rep003 SUCCESSFUL
[02.02|22:29:59] WARNING: Trying to obtain results of crashed or failed job ss_iter004_rep000
[02.02|22:29:59] WARNING: Trying to obtain results of crashed or failed job ss_iter004_rep000
[02.02|22:29:59] Obtaining results of ss_iter004_rep000 successful. However, no guarantee that they make sense
[02.02|22:29:59] Obtaining results of ss_iter004_rep000 successful. However, no guarantee that they make sense
[02.02|22:29:59] JOB plamsjob/ps_cond020/ss_iter004_rep000 Steady State Convergence: RESTART ABORTED
[02.02|22:29:59] JOB plamsjob/ps_cond020/ss_iter004_rep000 Steady State Convergence: JOB REMOVED
[02.02|22:29:59] JOB plamsjob/ps_cond020/ss_iter004_rep001 Steady State Convergence: JOB REMOVED
[02.02|22:29:59] JOB plamsjob/ps_cond020/ss_iter004_rep002 Steady State Convergence: JOB REMOVED
[02.02|22:29:59] JOB plamsjob/ps_cond020/ss_iter004_rep003 Steady State Convergence: JOB REMOVED
[02.02|22:30:00] JOB plamsjob/ps_cond020 FINISHED
[02.02|22:30:00] JOB plamsjob/ps_cond021 SUCCESSFUL
[02.02|22:30:00] JOB plamsjob/ps_cond020 SUCCESSFUL
[02.02|22:30:03] JOB plamsjob FINISHED
[02.02|22:30:14] JOB plamsjob SUCCESSFUL
If the execution got up to this point, everything worked as expected. Hooray!
Finally, in the following lines, we just nicely print the results in a
table. See the API documentation to learn more about how the results
object is structured, and the available methods. In this case, we use
the turnover_frequency() and average_coverage() methods to get
the TOF for the gas species and average coverage for the surface
species, respectively. Regarding the latter one, we use the last 10
steps in the simulation to calculate the average coverages. Notably, the
loop iterates over the internal indices (results.indices()) of each
condition. i is simply a sequential number for each condition in
this case, but idx is an id for the corresponding child job. This id
may be a complex object for n-dimension (n>1) scanning parameters, so if
you want to access the properties of one of the child jobs, we recommend
using a loop like the one we use here. In the lines that follow, we use
this idx to get the maximum time that the simulation required to
achieve the steady state for that specific composition (“max time”).
x_CO = []
ac_O = []
ac_CO = []
TOF_CO2 = []
max_time = []
results_dict = results.turnover_frequency()
results_dict = results.average_coverage( last=10, update=results_dict )
for i,idx in enumerate(results.indices()):
x_CO.append( results_dict[i]['x_CO'] )
ac_O.append( results_dict[i]['average_coverage']['O*'] )
ac_CO.append( results_dict[i]['average_coverage']['CO*'] )
TOF_CO2.append( results_dict[i]['turnover_frequency']['CO2'] )
max_time.append( results.children_results( child_id=idx ).history( pos=-1 )['max_time'] )
print( "-----------------------------------------------------------" )
print( "%4s"%"cond", "%8s"%"x_CO", "%10s"%"ac_O", "%10s"%"ac_CO", "%12s"%"TOF_CO2", "%10s"%"max_time" )
print( "-----------------------------------------------------------" )
for i in range(len(x_CO)):
print( "%4d"%i, "%8.2f"%x_CO[i], "%10.6f"%ac_O[i], "%10.6f"%ac_CO[i], "%12.6f"%TOF_CO2[i], "%10.3f"%max_time[i] )
-----------------------------------------------------------
cond x_CO ac_O ac_CO TOF_CO2 max_time
-----------------------------------------------------------
0 0.20 0.999250 0.000000 0.002567 20.000
1 0.21 0.999900 0.000000 0.000000 20.000
2 0.22 0.999590 0.000000 0.000700 20.000
3 0.23 0.999130 0.000000 0.002100 20.000
4 0.24 0.998630 0.000000 0.004267 20.000
5 0.25 0.998720 0.000000 0.003267 20.000
6 0.26 0.998760 0.000000 0.003267 20.000
7 0.27 0.998920 0.000000 0.003233 20.000
8 0.28 0.998090 0.000000 0.007100 20.000
9 0.29 1.000000 0.000000 0.003241 80.000
10 0.30 1.000000 0.000000 0.004973 80.000
11 0.31 1.000000 0.000000 0.005157 80.000
12 0.32 0.992570 0.000020 0.026933 20.000
13 0.33 1.000000 0.000000 0.007310 80.000
14 0.34 1.000000 0.000000 0.011559 80.000
15 0.35 1.000000 0.000000 0.016751 80.000
16 0.36 1.000000 0.000000 0.022879 80.000
17 0.37 1.000000 0.000000 0.041531 80.000
18 0.38 0.999260 0.000000 0.069112 80.000
19 0.39 0.985810 0.000140 0.146791 80.000
20 0.40 0.975090 0.000140 0.214842 320.000
21 0.41 0.905970 0.000780 0.422818 320.000
22 0.42 0.856420 0.001410 0.613742 80.000
23 0.43 0.806960 0.002770 0.795856 80.000
24 0.44 0.778850 0.003330 0.975367 20.000
25 0.45 0.739230 0.004710 1.117933 80.000
26 0.46 0.728010 0.005240 1.278163 80.000
27 0.47 0.688730 0.008080 1.455912 80.000
28 0.48 0.624570 0.013870 1.625328 80.000
29 0.49 0.607870 0.014540 1.831873 80.000
30 0.50 0.568050 0.019630 2.021464 80.000
31 0.51 0.532990 0.034100 2.234967 20.000
32 0.52 0.472360 0.050070 2.461300 20.000
33 0.53 0.270100 0.359930 2.502505 80.000
34 0.54 0.000000 1.000000 0.191716 80.000
35 0.55 0.000000 1.000000 0.066715 80.000
36 0.56 0.000000 1.000000 0.000000 20.000
37 0.57 0.000000 1.000000 0.000000 20.000
38 0.58 0.000000 1.000000 0.000000 20.000
39 0.59 0.000000 1.000000 0.000000 20.000
40 0.60 0.000000 1.000000 0.000000 20.000
41 0.61 0.000000 1.000000 0.000000 20.000
42 0.62 0.000000 1.000000 0.000000 20.000
43 0.63 0.000000 1.000000 0.000000 20.000
44 0.64 0.000000 1.000000 0.000000 20.000
45 0.65 0.000000 1.000000 0.000000 20.000
46 0.66 0.000000 1.000000 0.000000 20.000
47 0.67 0.000000 1.000000 -0.000000 20.000
48 0.68 0.000000 1.000000 0.000000 20.000
49 0.69 0.000000 1.000000 0.000000 20.000
50 0.70 0.000000 1.000000 0.000000 20.000
51 0.71 0.000000 1.000000 -0.000000 20.000
52 0.72 0.000000 1.000000 -0.000000 20.000
53 0.73 0.000000 1.000000 0.000000 20.000
54 0.74 0.000000 1.000000 0.000000 20.000
55 0.75 0.000000 1.000000 0.000000 20.000
56 0.76 0.000000 1.000000 0.000000 20.000
57 0.77 0.000000 1.000000 -0.000000 20.000
58 0.78 0.000000 1.000000 0.000000 20.000
59 0.79 0.000000 1.000000 0.000000 20.000
60 0.80 0.000000 1.000000 0.000000 20.000
The above results are the final aim of the calculation. However, we can take advantage of python libraries to visualize them. Here, we use matplotlib. Please check the matplotlib documentation for more details at matplotlib. The following lines of code allow visualizing the effect of changing the \(CO\) molar fraction on the average coverage of \(O*\) and \(CO*\) and the production rate of \(CO_2\):
import matplotlib.pyplot as plt
fig = plt.figure()
ax = plt.axes()
ax.set_xlabel('Molar fraction CO', fontsize=14)
ax.set_ylabel("Coverage Fraction (%)", color="blue", fontsize=14)
ax.plot(x_CO, ac_O, color="blue", linestyle="-.", lw=2, zorder=1)
ax.plot(x_CO, ac_CO, color="blue", linestyle="-", lw=2, zorder=2)
plt.text(0.3, 0.9, 'O', fontsize=18, color="blue")
plt.text(0.7, 0.9, 'CO', fontsize=18, color="blue")
ax2 = ax.twinx()
ax2.set_ylabel("TOF (mol/s/site)",color="red", fontsize=14)
ax2.plot(x_CO, TOF_CO2, color="red", lw=2, zorder=5)
plt.text(0.37, 1.5, 'CO$_2$', fontsize=18, color="red")
plt.show()
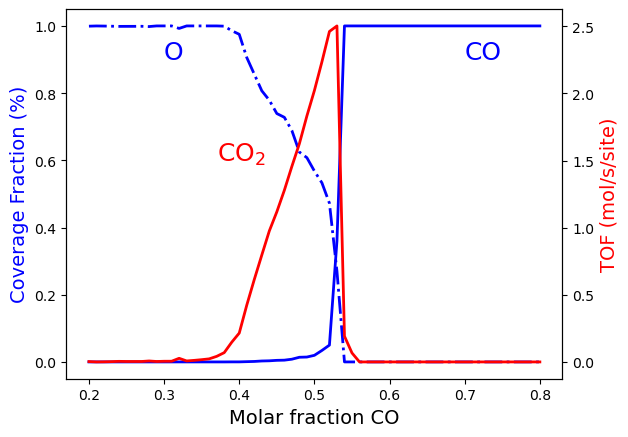
As shown in the Figure, we also found three regions involving two phase
transitions, which are similar to the ones obtained in the example
Phase Transitions in the ZGB model, where we did not converge the
calculation up to the steady-state condition. However, in this case, the
first transition, corresponding to the limit of \(O^*\) poisoning,
occurs at x_CO=0.40 rather than x_CO=0.32, and the second
transition, corresponding to the beginning of CO poisoning, occurs
roughly at the same value x_CO=0.55 but as a more abrupt change.
Now, we can close the pyZacros environment:
scm.pyzacros.finish()
[02.02|22:31:38] PLAMS run finished. Goodbye