Rationalize your Chemical Reactions
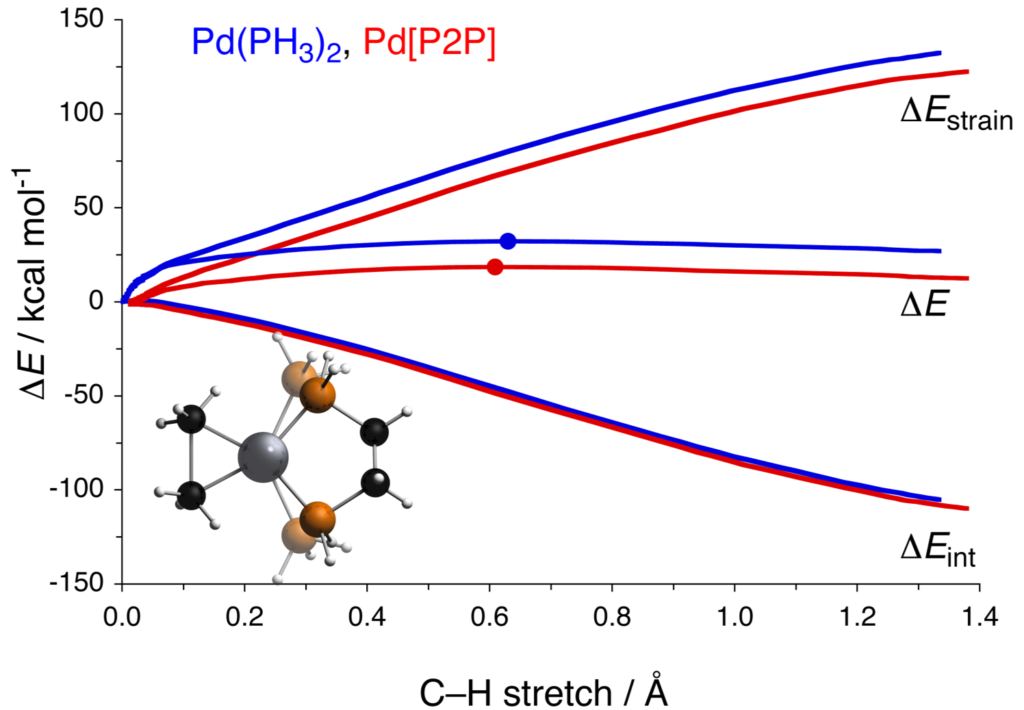
A recent Nature protocol paper describes a, step-by-step, easy to follow protocol to assist scientists with rationalizing factors controlling molecular reactivity [1]. Our close collaborators from the theoretical chemistry department at the VU describe how their activation strain model (ASM) [2] decomposes the potential energy along a reaction coordinate into the reaction strain, or distortion energy, and the interaction energy between increasingly distorted reactants.
The strain energy is the energy needed to deform the reactants from their equilibrium geometry into the geometries they acquire to react and the interaction energy is the actual chemical interaction between the two deformed reactants. There are robust off-the-shelf-solutions that permit researchers to calculate barrier heights and look into geometries of transition states.
ADF can bring value to this protocol with its energy decomposition analysis (EDA) scheme, in which the interaction energy between the reactants is even further decomposed into a number of physically meaningful terms such as the classical electrostatic interaction, steric (Pauli) repulsion, and the stabilizing orbital interactions. This means the experimentalist can obtain in-depth information on the type of interaction and hence, rationalise the driving factors of their reaction in more detail.
The protocol effectively equips chemists to first rationalize in silico whether a particular reaction is strain- or interaction-controlled. They can then build on this quantitative insight to further tune the reaction and remove the unwanted and often employed trial-and-error approach to methodology development. Researchers empowered with these methods can save time and chemical waste by reducing the number of experiments, focusing only on the most promising catalysts and reaction conditions.
While the protocol works most naturally with ADF, other codes can also be used. Suggestions for DFT functionals and basis sets are provided and the supplementary materials to this work comprise a selection of input files which will ultimately streamline the learning process to adopt this approach to the particular chemistry under study.
[1] Vermeeren, P., van der Lubbe, S.C.C., Fonseca Guerra, C. et al. Understanding chemical reactivity using the activation strain model. Nat. Protoc. 15, 649–667 (2020).
[2] F. M. Bickelhaupt, K. N. Houk, Analyzing Reaction Rates with the Distortion/Interaction‐Activation Strain Model, Angew. Chem. Int. Ed. 56, 10070 (2017).
Key conceptsADF bonding analysis catalysis Reactivity