Tips and Tricks for Transition State Searches for Click Reactions¶
Contents¶
This tutorial will guide you through more a complicated transition state search where the reactants do not approach from the same plane. The transition state for the Pyrithione tautomerization in the AMS-Tutorials
was rather simple as the hydrogen atom moves along the O…S direction and the transition state (TS) can be expected somewhere in between. A Potential Energy Surface (PES) scan along the H…S distance can help to find a maximum in energy which can serve as input for a TS calculation. You may want to revisit this tutorial if you are not familiar with PES scans.
Some reactions are more complicated and have more complex moves which do not easily translate to a simple PES coordinate scan. So how can we get a good guess geometry?
- Find an Organic Textbook of your choice that discusses reaction mechanisms.
- Collaborate with an Organic Chemist.
- Use a molecule kit and build the structures you need and act out possible mechanisms.
We will use the Diels-Alder reaction to explain the thought processes of a modeller for click reactions. The Diels-Alder reaction mechanism is a click reaction to build six membered rings from scratch using a diene and a dienophile. The simplest six membered ring can be formed with a molecule of butadiene as diene and a molecule of ethene as dienophile which react to form Cyclo-1-hexene.
The reaction above is presented correctly but simplified. If you study the literature you will find out that
- Diene and Dienophile are not approaching each other in the same plane.
- Cyclo-1-hexene is not flat.
As Cyclo-1-hexene is non-aromatic and has only one double bond we expect the ring to be more a distorted chair than a planar ring. Equipped with this information we will use the ADF GUI and start our work from there.
Tools¶
We will work with ADF and DFTB. We will use routines for optimisations, frequency calculations, Potential Energy Surface (PES) scans, and Nudge Elastic Band (NEB).
Pre-requisites¶
You should be proficient to comfortably draw a structure, set up a calculation when given hints, run it and retrieve results. You may want to refresh your skill in the GUI tutorials
Setting up the Structure¶
There are many ways how diene and dienophile can approach each other – too many to try them all. We are, however, very certain what the result of our reaction looks like, so we will start from there.
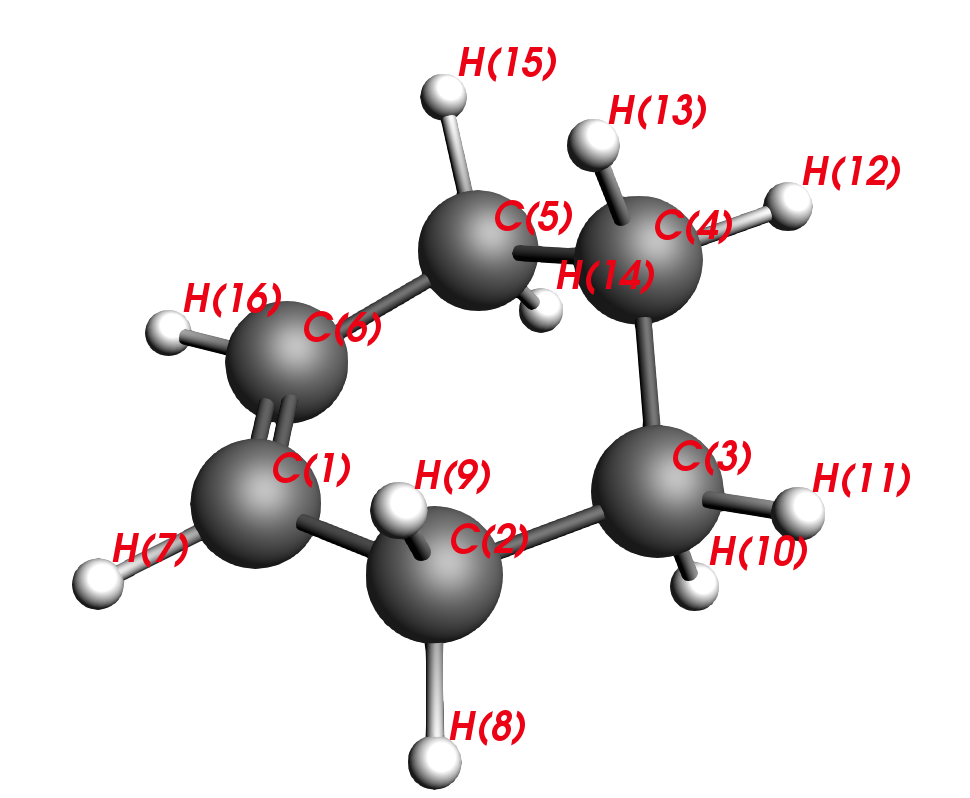
- Open ADFinputUse the drawing tools create a molecule of Cyclo-1-hexeneMake sure it’s not planar and pull one of the carbon atoms out of the plane
Note
The algorithm to find a minimum in ADF is programmed in such a way to find the nearest local minimum. By forcing the ring to be planar, indeed, a minimum can be found. If you perform frequency calculations on this planar structure, you would notice that there are 2 small negative frequencies – and you are stuck on some saddle point of the second order. Making the structure aplanar opens a new search space and you will find a true local minimum with no negative frequencies.
If you are uncertain how to manipulate, you can use a start structure from here
Setting up the calculations¶
After manipulating our input to an aplanar structure we will perform an optimization to find a minimum for Cyclo-1-hexene.
- From the Main Tab choseTask → Geometry OptimizationXC-Functional → GGA-D → PBE-D3(BJ)Relativity → noneBasis set → TZPFrozen Core → noneNumerical Quality → Normal
Note
These settings will provide you with a good starting point for organic reactions in general.
- Save the file as DA_product.adf and run the calculation
When you look at the output split the molecule in your mind to the reaction partners that must have formed this product.
C2-C1-C6-C5 are carbons that stem from the diene and C3-C4 are carbons that stem from the dienophile. Thus, our next task will be to reverse the reaction and move the reactants apart, similar to unclicking of LEGO bars. This is the intuitive approach to finding a transition state.
Follow your intuition¶
Starting from the optimised structure from Chapter 2, we will setup a PES scan and we will use a DFTB method to speed up the process.
- Switch from ADF to DFTBIn the Main tab choseTask → PES ScanModel → SCC-DFTBParameter directory → DFTB.org/3ob-3-1
- Select the C2 and C3 atoms in the GUI using your left mouse button.Press + to add the scan coordinate.Set Number of scan points for coordinate to
10.Set the scan range from1.53to3Å.Save the file asDA_PES_DFTB.adfand run the calculation.
The calculation may finish with a warning such as:
This is ok for us as we are just after a good intuitive guess, so we can ignore this message.
- Select SCM → Movie.Click on the maximum point of the red curve.
You will be presented with the corresponding structure and you can recognize already the diene in one plane and the dienophile approaching from above, as described in a textbook for this type of reaction. We will use this structure to perform a frequency calculation to see where on the PES we actually are and to check if this structure is suitable for a transition state (TS) calculation.
- In ADFMovie, using either arrow keys or the slider, select Frame number 6 (or the frame where the maximum is)Click on File → Update Geometry in InputThis will bring ADFInput to the front updated with the geometry of Frame 6
Performing a TS calculation is not simple; we are looking for a special point of the PES, a saddle point of the first order. This is where the curvature of the PES area is positive in all directions but one. In other words, we are looking for one negative frequency that corresponds to the move along our reaction coordinates, i.e. C2-C3 and C5-C6.
Thus, we will have to do a frequency calculation.
- In ADFInput, go to the Main panelSelect Task → Single PointSelect Followed by → Frequencies
We can now run the Frequency calculation, which will compute the Hessian and the normal modes:
- Click on File → Save As... and name it
DA_Freq_DFTB.adf.Submit the calculation and wait for it to finish.Then click on SCM → Spectra
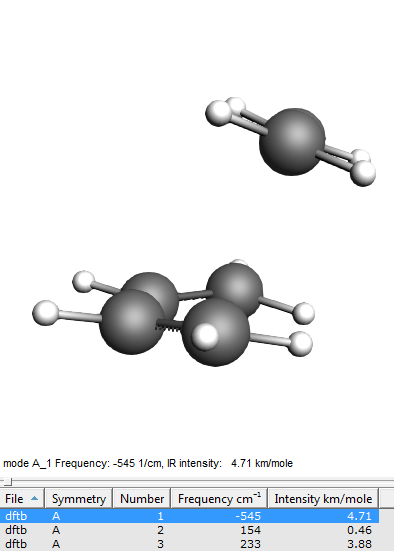
The first mode should have an imaginary frequency (which in the table is shown as a negative frequency). Click on the line in the table corresponding to the imaginary frequency to visualize the mode. You will see C2 and C3 moving towards and away from each other as you would expect them to do in the actual reaction.
Now we are ready to let DFTB find the real TS.
- Open the ADFInput window of the job “DA_Freq_DFTB”Select Task → Transition StateMake sure that Followed by is set to Frequencies
You will need the frequency file to assist the TS calculation. The algorithm needs the Hessian as input.
- Go to the panel Details → Geometry OptimizationSelect Initial Hessian → From FileClick on the folder icon next to Initial Hessian From:Select the file dftb.rkf in the folder DA_Freg_DFTB.results
Note that your input still carries the geometry constraint from the previous PES scan calculation, which has to be removed before starting the new calculation:
- Go to the panel Model → Geometry Constraints and PES ScanRemove the C(2) C(3) constraint by clicking on “-“ in front of it.
We can now run the TS search calculation:
- Click on File → Save As... and give it the name DA_TS_DFTB.adfSubmit the calculation and wait for it to finish.Then click on SCM → Spectra
Note
There is only ONE negative frequency defining a transition state!
You will see to molecules, clearly identifiable as butadiene and ethene, only one negative frequency, and when you animate the vibration you see the molecules moving towards each other as if they would form the product and away from each other, as if they were splitting up again.
What can possibly go wrong?¶
You could find more than one negative frequency which means you are lost on the PES and have not found the TS. This can happen if the PES scan goes wrong because you did not have a good starting structure. In the case of click reactions, rearrange the two reactants in such a fashion that the resemble the suggested text book mechanism, i.e.
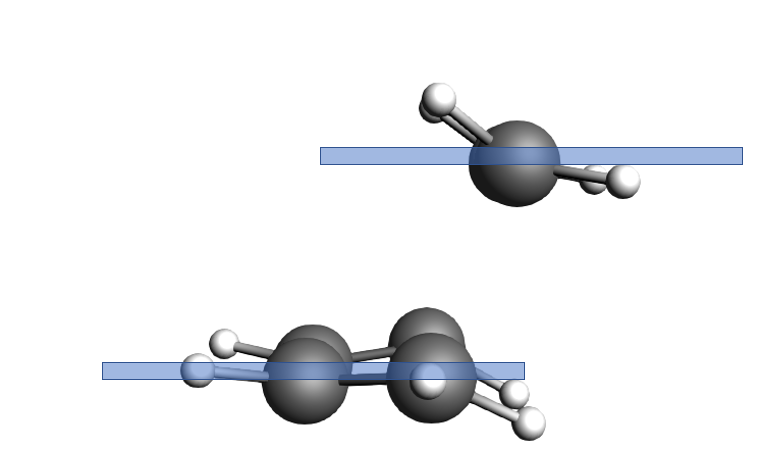
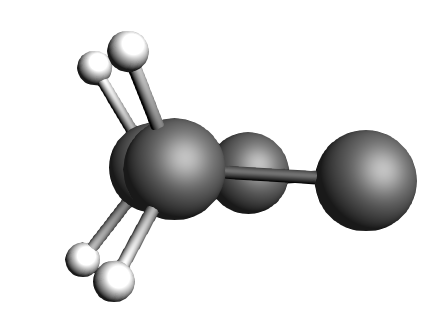
Fig. 3 The ethene molecule is in one plane and the butadiene in another one, very parallel to each other.

Fig. 4 The ethene molecule hovers “perfectly” above the butadiene C atoms it is supposed to click with.
… then repeat the PES.
Animate the negative frequencies and identify the one that represents the reaction coordinate, i.e. the one that looks like a movie of the reaction to happen. Then animate the other negative ones that have nothing to do with the reaction. Some common scenarios can be:
| Problem | Solution |
|---|---|
| geometries are forced to be planar, but should not | force a slight aplanarity, repeat TS calculation |
| local geometries are staggered, but should be eclipsed | change the torsion angle, repeat TS calculation |
What are the next steps?¶
- Reuse the DFTB geometry and Hessian to calculate the TS at the DFT level, e.g. PBE-D3(BJ)/TZP. This works well if the DFT and DFTB potential energy surfaces are reasonably similar. Switch to ADF with the updated geometry, select Transition State as Task. In the Details → Geometry Convergence panel click on the folder next to the Initial Hessian field and select the dftb.rkf from the DFTB TS calculation.
- If the TS does not converge from the DFTB guess, you could retry the PES scan and initial Hessian calculation with ADF.
- After you’ve obtained a TS at the DFT level, perform a frequency calculation again to confirm that there is really only one negative frequency, and thus you are confident about your calculation.
- Report geometries and frequency values in your paper (supporting information).
- Perform an IRC search to find minima of reactants and products, see here
Starting from reactant and product – Nudge Elastic Band (NEB)s¶
Another way to locate a TS would be using the NEB method. This requires you to provide the structure of your reactants and your products. These structures you provide have to have a realistic geometry but do not necessarily have to be minima on the PES. In case of our Diels-Alder reaction we do know about the product but we are uncertain how exactly the diene and dienophile look in their local minima before they “started to click”.
The NEB algorithm does not know about chemical reactions; it will interpolate between the geometries of your reactants and products. A preliminary path on the PES can be established which will then be discretized into a finite number of points. Each point corresponds to a geometry which the reactants are adopting on their way to reach the product. Imagine you would thread these images like pearls on a necklace, but using a rubber band. The NEB algorithm will check the slope of the PES and “nudges” the rubber band toward the minimum energy path (MEP). The MEP is defined as the path on the PES where every point is at an energy minimum in all directions perpendicular to the path. It also passes through at least one first-order saddle point which is the transition state we are looking for.
To help you select two suitable input structures, let’s revisit your file DA_PES_DFTB.adf.
- Open the file as DA_PES_DFTB.adf and from the SCM Menu choose Movie.Click on the Frame 0 of the red curve
- Choose File → Save Geometry... and save the file as
product.xyz
As our reactant, we could use frame 7 in our results.
- Click on the Frame 7 of the red curveChoose File → Save Geometry... and save the file as reactants.xyz
We now need to load both files into the ADF GUI.
- Start a new ADFinputClick on File → Import coordinates... and select the file reactant.xyzAdd a new molecule: Edit → New MoleculeClick on File → Import coordinates... and select the file product.xyz
You are now ready to setup your calculation:
- In the Main tab chose | Task → Transition StateXC-Functional → GGA-D → PBE-D3(BJ)Relativity → noneBasis → TZPFrozen Core → NoneNumerical Quality → Normal
Select the details for the Transition State search:
Note
We chose 3 to speed up the tutorial. In research conditions, use the default (8) or bigger.
- Click on File → Save As... and give it the name DA_NEB.adf.Submit the calculation and wait for it to finish.Then click on SCM → Movie
The highest point in energy gives you a good starting structure for a TS calculation.
If you play the movie you should see the reaction happen.
What can possibly go wrong?¶
You could fail with the NEB and find no TS. In this case revisit the structure you are least sure – which would be the reactants in our case. Imagine again each point on the NEB preliminary path corresponds to a geometry which the reactants are adopting on their way to reach the product. Make a movie out of it in your mind’s eye. Does the reaction look smooth? Are atoms forced to penetrate each other to change the geometry from reactant to product? If so, correct the geometries in such a way that this does not happen and start the NEB afresh.
What are the next steps?¶
- Use the TS obtained with DFTB and perform a frequency calculation using ADF with the settings from before, i.e. PBE-D3(BJ) with a TZP basis set to get a guess for the Hessian. (initial guess for TS calculation)
- Use the Hessian to perform a TS calculation. (actual TS calculation)
- Do perform a frequency calculation again to confirm that there is really only one negative frequency, and thus you are confident about your calculation. (confirmation of TS)
- Report geometry and frequency values in your paper.
- Perform an IRC search to find minima of reactants and products, see here