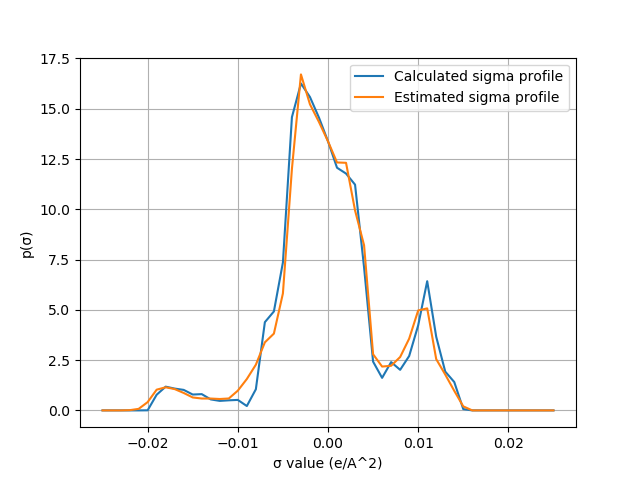
Calculating and estimating sigma profiles¶
Sigma profiles are one of the fundamental pieces of a COSMO-RS/-SAC calculation. They are also widely used as an important empirical descriptor for a molecule’s behavior in a solution as well as for a molecule’s properties in a number of applications. In the standard COSMO-RS/-SAC workflow, sigma profiles are generated after a sequence of DFT calculations which – for large molecular systems – can take considerable time to complete. For computationally expensive systems or high-throughput screening applications, it is sometimes advantageous to approximate sigma profiles using tools like fast_sigma from AMS.
In the following python script, we generate sigma profiles for n-Hexanoic acid using the two approaches discussed above. The function calc_sigma_profile will generate sigma profiles from .coskf files, and the function fast_sigma will generate sigma profiles from SMILES strings using the fast_sigma tool.
Python code¶
[show/hide code]
import os
import numpy as np
import matplotlib.pyplot as plt
from scm.utils.runsubprocess import RunSubprocess
from scm.plams import Settings, init, finish, CRSJob, config, KFFile
import subprocess
######## Note: Ensure to configure the database path to either the installed ADFCRS-2018 directory or your own specified directory ########
database_path = os.path.join(os.environ["SCM_PKG_ADFCRSDIR"], "ADFCRS-2018")
if not os.path.exists(database_path):
raise OSError(f"The provided path does not exist. Exiting.")
init()
# suppress plams output
config.log.stdout = 0
class SigmaProfile:
def __init__(self, chdens, profiles, profile_names):
if len(profiles) != len(profile_names):
print("Error: profiles_names and profiles of different sizes")
self.chdens = chdens.flatten() if isinstance(chdens, np.ndarray) else chdens
self.profiles = {
name: prof.flatten() if isinstance(prof, np.ndarray) else prof
for name, prof in zip(profile_names, profiles)
}
def __str__(self):
line = "─" * (15 * (1 + len(self.profiles)))
ret = (
line
+ "\n"
+ "".join(["Charge Dens.".ljust(15)] + [name.ljust(15) for name in self.profiles])
+ "\n"
+ line
+ "\n"
)
for i in range(len(self.chdens)):
ret += "{0:.5g}".format(self.chdens[i]).ljust(15) + "".join(
["{0:.5g}".format(v[i]).ljust(15) for k, v in self.profiles.items()]
)
ret += "\n"
return ret
def fast_sigma(smiles):
results_file = "tmp_results18954.compkf"
subprocess_string = " --smiles '" + smiles + "'"
if not os.path.isfile(os.path.join(os.path.expandvars("$AMSBIN"), "fast_sigma")):
raise OSError("ERROR: cannot find fast_sigma ... has amsbashrc been executed?")
fs = os.path.join(os.path.expandvars("$AMSBIN"), "fast_sigma")
scm_sp = RunSubprocess(fs + subprocess_string + " -o " + results_file)
if os.path.isfile(results_file):
crskf = KFFile(results_file)
res = crskf.read_section("PURESIGMAPROFILE")
sp = SigmaProfile(
chdens=res["chdval"],
profiles=[res["profil"], res["hbprofil"]],
profile_names=["total_profile", "HB_profile"],
)
os.remove(results_file)
return sp, scm_sp
else:
return None, scm_sp
def calc_sigma_profile(coskf_file, cosmosac=False):
# initialize settings object
settings = Settings()
settings.input.property._h = "SIGMAPROFILE"
# set the number of compounds
compounds = [Settings()]
compounds[0]._h = os.path.join(database_path, coskf_file)
compounds[0].frac1 = 1
# to change to the COSMOSAC2013 method
if cosmosac:
settings.input.method = "COSMOSAC2013"
# specify the compounds as the compounds to be used in the calculation
settings.input.compound = compounds
# create a job that can be run by COSMO-RS
my_job = CRSJob(settings=settings)
out = my_job.run()
res = out.get_results()
if cosmosac:
prof_len = len(res["hbprofil"]) // 3
sp = SigmaProfile(
chdens=res["chdval"],
profiles=[res["profil"]] + [res["hbprofil"][i * prof_len : (i + 1) * prof_len] for i in range(3)],
profile_names=["total_profile", "HB", "HB-OH", "HB-OT"],
)
else:
sp = SigmaProfile(
chdens=res["chdval"],
profiles=[res["profil"], res["hbprofil"]],
profile_names=["total_profile", "HB_profile"],
)
return sp
# regular way to generate a sigma profile from a .coskf file
filename = "n-Hexanoic_acid.coskf"
sp = calc_sigma_profile(filename, cosmosac=False)
# way using the fast_sigma estimation method
fs_sp, err = fast_sigma("CCCCCC(=O)O")
if fs_sp is None or len(err[1]) > 0:
print("fast_sigma generated the following output:\n" + err[1])
plt.xlabel("σ value (e/A^2)")
plt.ylabel("p(σ)")
plt.plot(sp.chdens, sp.profiles["total_profile"], label="Calculated sigma profile")
if fs_sp is not None:
plt.plot(fs_sp.chdens, fs_sp.profiles["total_profile"], label="Estimated sigma profile")
plt.legend(loc="upper right")
plt.grid()
# plt.savefig(f"./Sigma_profile.png")
plt.show()
finish()
This code produces the following output: