Eutectic systems¶
A eutectic point of a chemical mixture defines the minimum melting composition of that system over the composition range. In other words, the eutectic point will have a lower melting point than the pure components making up the mixture as well any other possible mixture. In this example, we calculate the eutectic point of a binary mixture of ethanol and water as the intersection of the solid-liquid equilibrium curves of two systems: (1) Solid ethanol dissolved in water and (2) Solid water dissolved in ethanol. This script will output the mole fraction of ethanol at the eutectic point as well as the temperature. For comparison, Takaizumi and Wakabayashi [1] provide an experimental eutectic point with a mole fraction value of 0.86 for ethanol and a melting temperature of -124.3 °C.
Python code¶
[show/hide code]
import os
import matplotlib.pyplot as plt
from scm.plams import Settings, init, finish, CRSJob, config
######## Note: Ensure to configure the database path to either the installed ADFCRS-2018 directory or your own specified directory ########
database_path = os.path.join(os.environ["SCM_PKG_ADFCRSDIR"], "ADFCRS-2018")
if not os.path.exists(database_path):
raise OSError(f"The provided path does not exist. Exiting.")
init()
# suppress plams output
config.log.stdout = 0
# the ethanol water system
# experimental numbers for this eutectic are 0.86 mole fraction ethanol at a temperature of -124.3 degrees C
files = ["Ethanol.coskf", "Water.coskf"]
tm = [158.5, 273.15] # K
hfus = [1.2, 1.43] # kcal/mol
initial_t_range = [100, 300] # K -- the temperature range over which the eutectic search is done
steps = 20 # number of steps to take within the temperature range
# another eutectic system
# files = ["L-Menthol.coskf","Camphor.coskf"]
# tm = [316.2,451.5] #K
# hfus = [2.84,1.63] #kcal/mol
# initial_t_range = [100,460] #K
# steps = 20
# if we know the eutectic temperature is bounded to within a range of <= estimate_precision, we simply use a linear interpolation between two x,T pairs
estimate_precision = 1.0 # K
class Eutectic:
def __init__(self, files, tm, hfus, steps=10):
self.files = files
self.tm = tm
self.hfus = hfus
self.steps = steps
if not (len(self.files) == len(self.tm) == len(self.hfus)):
print("Error. Inputs must be the same length.")
def calc_xt_curves(self, t_range):
# initialize settings object
settings = Settings()
settings.input.property._h = "SOLUBILITY"
# optionally, change to the COSMOSAC2013 method
settings.input.method = "COSMOSAC2013"
# make compounds
compounds = [Settings() for i in range(len(files))]
for i, file in enumerate(self.files):
compounds[i]._h = os.path.join(database_path, file)
compounds[i].meltingpoint = self.tm[i]
compounds[i].hfusion = self.hfus[i]
comp1_fracs = []
for i, frac1 in enumerate([0.0, 1.0]):
compounds[0].frac1 = frac1
compounds[1].frac1 = 1.0 - frac1
settings.input.temperature = " ".join([str(t) for t in t_range] + [str(self.steps)])
# add the compounds to the settings object
settings.input.compound = compounds
# create a job that can be run by COSMO-RS
my_job = CRSJob(settings=settings)
# run the job
out = my_job.run()
# convert all the results into a python dict
res = out.get_results()
if i == 0:
comp1_fracs.append(res["molar fraction"][0])
else:
comp1_fracs.append(1.0 - res["molar fraction"][1])
return comp1_fracs
def calc_eutectic(self, t_range, history=[[], [], []]):
comp1_fracs = self.calc_xt_curves(t_range)
# the temperatures used in the calculation
temps = [t_range[0] + (t_range[1] - t_range[0]) / self.steps * i for i in range(self.steps + 1)]
history[0].extend(comp1_fracs[0])
history[1].extend(comp1_fracs[1])
history[2].extend(temps)
# the difference between compound 1's mole fraction in the two calculations
# when these mole fractions are the same, we've found the eutectic
diffs = comp1_fracs[0] - comp1_fracs[1]
# find where the sign changes (intersection of SLE lines)
for i in range(self.steps):
if diffs[i] * diffs[i + 1] < 0:
if temps[i + 1] - temps[i] < estimate_precision:
# use linear combination of t's
tot = abs(diffs[i]) + abs(diffs[i + 1])
w1 = tot - abs(diffs[i]) # same as abs(diffs[i+1])
w2 = tot - abs(diffs[i + 1])
return (
(
(w1 * comp1_fracs[0][i] + w2 * comp1_fracs[1][i + 1]) / tot,
(w1 * temps[i] + w2 * temps[i + 1]) / tot,
)
), history
else:
return self.calc_eutectic([temps[i], temps[i + 1]], history)
return None, None
eutectic_calc = Eutectic(files, tm, hfus, steps=steps)
eutectic, history = eutectic_calc.calc_eutectic(initial_t_range)
if not eutectic:
print("No eutectic point found in the temperature range")
else:
print("Found eutectic point:")
print("x_1".rjust(10), "T (K)".rjust(10), "T (C)".rjust(10))
x, t = eutectic
print(str(x.round(5)).rjust(10), str(t.round(5)).rjust(10), str((-273.15 + t).round(5)).rjust(10))
# plot the solubility curves and eutectic point
h_s1 = sorted(list(zip(history[0], history[2])), key=lambda x: x[0])
h_s2 = sorted(list(zip(history[1], history[2])), key=lambda x: x[0])
h_s1 = [x for x in h_s1 if x[0] >= eutectic[0] and x[1] >= eutectic[1]]
h_s2 = [x for x in h_s2 if x[0] <= eutectic[0] and x[1] >= eutectic[1]]
# adjust the melting point back to the correct value for high or low solubility
for i in range(len(h_s1)):
if h_s1[i][0] > 0.9999:
h_s1[i] = (h_s1[i][0], tm[0])
for i in range(len(h_s2)):
if h_s2[i][0] < 0.0001:
h_s2[i] = (h_s2[i][0], tm[1])
plt.plot([x[0] for x in h_s1], [x[1] for x in h_s1], label="x_1 (solvent compound 1)")
plt.plot([x[0] for x in h_s2], [x[1] for x in h_s2], label="x_1 (solvent compound 2)")
plt.plot(eutectic[0], eutectic[1], "o", label="Eutectic point")
if eutectic[0] < 0.5:
plt.annotate(" " + str(tuple([xt.round(3) for xt in eutectic])), eutectic, va="center", ha="left")
else:
plt.annotate(str(tuple([xt.round(3) for xt in eutectic])) + " ", eutectic, va="center", ha="right")
plt.xlabel("Mole fraction compound 1")
plt.ylabel("Melting point of mixture (K)")
plt.legend(loc="upper right")
plt.grid()
plt.show()
finish()
This figure (produced by the code) shows the two solubility curves calculated by the program.
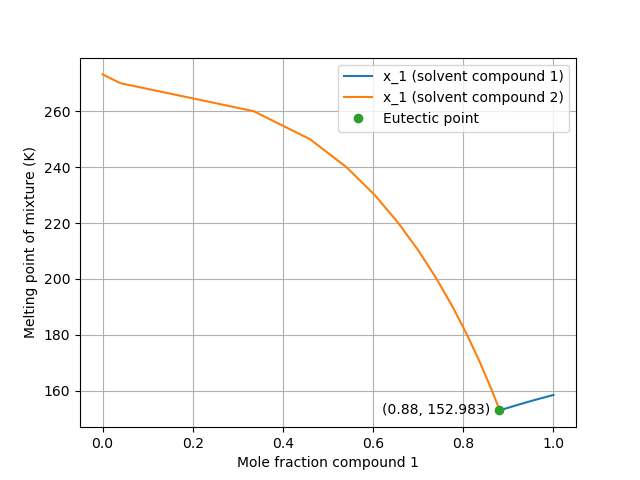
Found eutectic point:
x_1 T (K) T (C)
0.88028 152.98252 -120.16748