5.5. Parameter sensitivity analysis¶
Important
This tutorial is only compatible with ParAMS 2023.1 or later.
This example builds on ReaxFF (basic): H₂O bond scan. That example focuses on the optimization, this one focuses on the parameter sensitivity analysis.
When reparameterizing ReaxFF it is difficult to know which parameters to optimize.
Too few parameters might make it difficult to find a good fit.
Too many parameters will make the optimization harder, slower, and increase the risk of overfitting.
In this tutorial we will show you how to identify the most sensitive parameters which can guide your parameter selection.
The relevant files for this tutorial can be found in $AMSHOME/scripting/scm/params/examples/ReaxFF_water.
See also
5.5.1. How sensitivity is calculated¶
Sensitivity is calculated using the Hilbert-Schmidt Independence Criteria (HSIC), a robust and popular kernel-based dependence measure.
The HSIC equals zero if and only if variables are independent
Larger values imply higher dependency between variables
We will test the dependency of the loss function on each of the parameters we might consider optimizing.
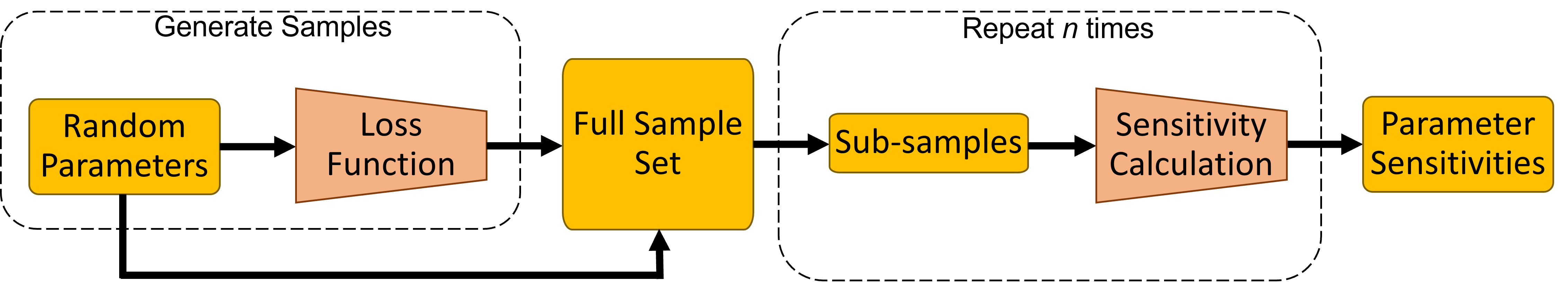
Fig. 5.7 Simplified sensitivity workflow schematic¶
The sensitivity calculation is performed as follows:
Select all the parameters you would like to investigate
Draw uniform random samples of parameters values
Calculate loss values for each of the parameter sets
For each parameter calculate the HSIC metric using the parameter values and loss function values
Calculate sensitivity values based on the HSIC metric
Sensitivity values have two properties which make them very easy to interpret:
all sensitivity values sum to one;
a sensitivity value ranges from 0 (insensitive) to 1 (sensitive);
5.5.2. Setting up a sensitivity calculation¶
First load the example files:
sensitivity.in.This will load the sensitivity calculation inputs and the YAML files.
Warning
Do not select another file when opening since this will default to the input file called params.in which is the
setup for the optimization version of this tutorial.
5.5.2.1. Selecting parameters¶
Next we consider the parameters we would like to investigate.
This shows that all 12 parameters related to an H.O bond are active.
If we consider the parameter descriptions we see that 6 of them are associated with π-bonds which are not present in the training set, so they should have no influence on the optimization.
Instead of deactivating these parameters, we will leave these parameters active as a test of the sensitivity method.
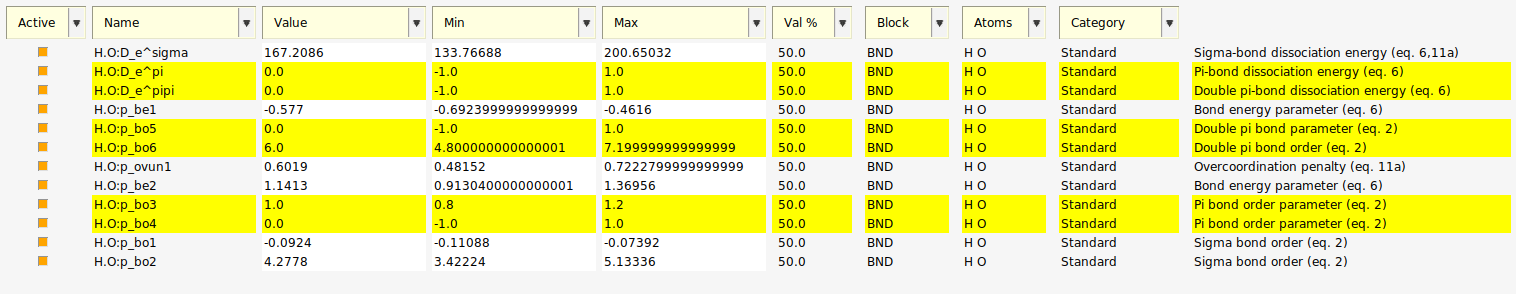
Fig. 5.8 12 active parameters with parameters related to π-bonds selected.¶
5.5.2.2. Input options¶
We will now configure the sensitivity calculation.
 to open the input panel
to open the input panel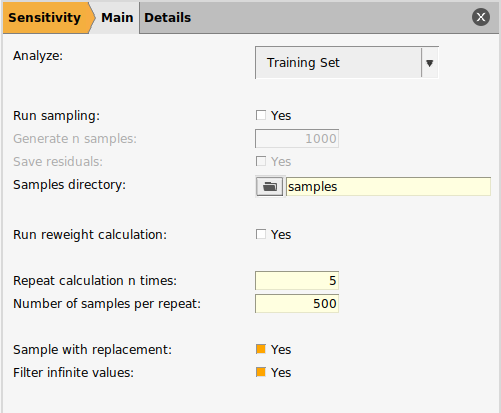
5.5.2.2.1. Set To Analyze¶
The Analyze option allows you to select if you would like to test the sensitivity of the training set or the validation set.
Investigating both can be very helpful in understanding why the two perform differently.
In this tutorial, we have no validation set and thus test only the training set.
5.5.2.2.2. Loading/Generating Samples¶
The second set of options relate to producing or loading the samples of the loss function. If you have produced samples before, you can use them again for a different sensitivity calculation to save time because sampling takes longer than the sensitivity calculation.
In this tutorial you can choose to either
Load samples from the sample directory (to get through the tutorial quicker), or
Generate the samples yourself (if no samples had been generated previously, this would be a necessary step)
 and select the
and select the samples directory in the example directoryThe associated input keys for loading samples:
RunSampling No
SamplesDirectory path/to/samples
In general the Samples Directory must have the following structure:
samples_directory
├── training_set_results (or validation_set_results)
│ ├── running_active_parameters.txt
│ └── running_loss.txt
└── glompo_log.h5
Only
glompo_log.h5or therunning_*.txtfiles are needed.If
glompo_log.h5is present, it will take precedence.glompo_log.h5is produced by saving residuals (see the To generate samples tab).
Saving residuals gives you:
the difference between prediction and reference value for every item in your training set for every parameter set evaluated;
detailed insight into your training set;
necessary data for the reweight calculation;
an HDF5 log file which can take a lot of disk space.
We do not require the residuals for the calculation, so we will not save them.
The input keys to generate your own samples:
RunSampling Yes
SaveResiduals No
NumberSamples 2000
Tip
Sampling will produce a directory called sampling_results within the results directory.
To load these samples point Samples Directory to: results/sampling_results/optimization.
5.5.2.2.3. Samples used in calculation¶
Our pool of samples contains 2000 samples. To take full advantage of these we can:
Use all 2000 samples in a single calculation; or
Take a subset of the samples, run a smaller calculation, and repeat this for different subsets.
It is always recommended to repeat the sensitivity calculation. The spread of results will tell you if you have enough samples. It is usually cheaper to repeat a calculation with subsets of samples than it is to run it using all samples once.
We will repeat the calculation five times using subsets of 500 samples each time.
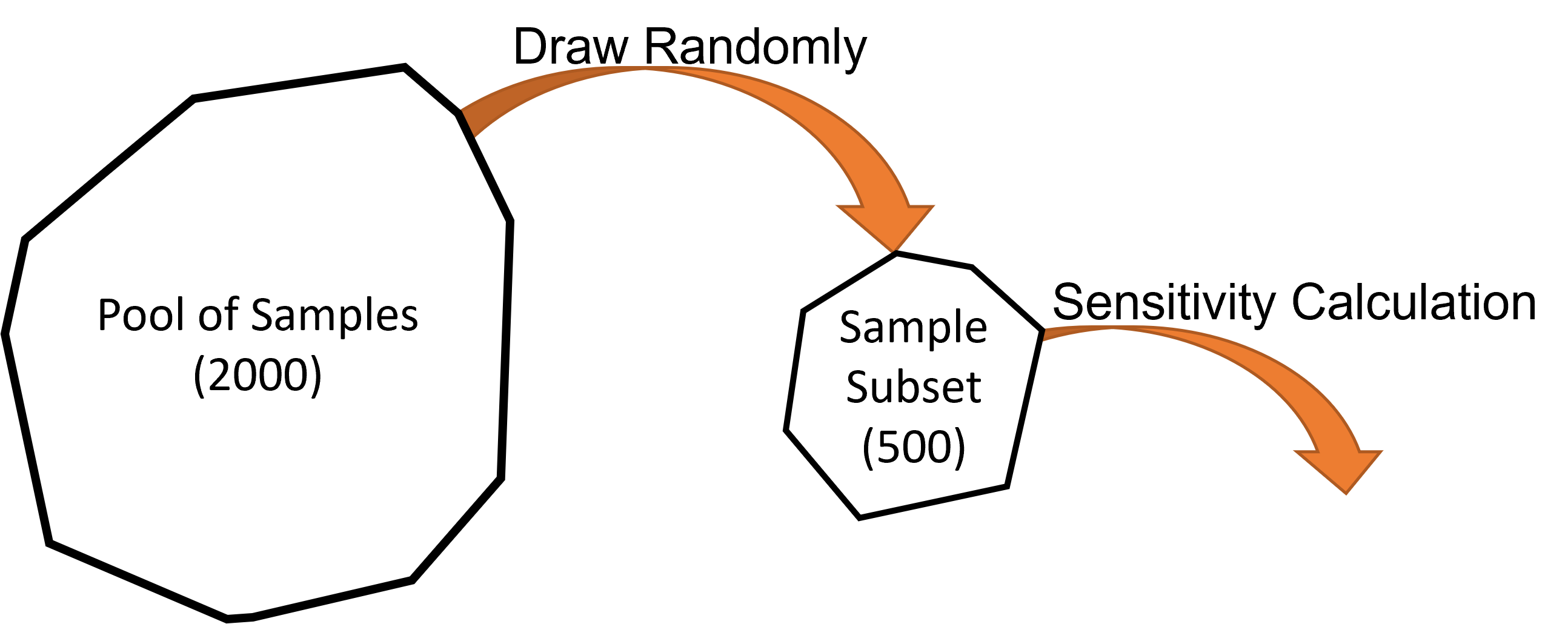
Fig. 5.9 The sensitivity calculation is run on a subset of the pool of samples.¶
A new subset is drawn from the full pool for every calculation repeat.
If Sample with replacement is Yes then the subset can contain repeated samples from the full pool of samples. Otherwise, a sample can only appear once in a subset.
If Filter infinite values is Yes then any samples with an infinite loss function value will not be included in the full pool of samples.
Tip
If Run Reweight Calculation is Yes, then the sensitivity calculation is extended to provide suggested training set weights which should provide a better balance of sensitivities for the parameter set. This training set only has one item so we will not run the reweight calculation.
Data sets which are imbalanced can be harder to optimize. A reweight calculation
suggests new training set weights,
reveals dependencies within the training set,
can help to guide training set design.
Unfortunately, a reweight calculation is much more computationally demanding than a regular sensitivity calculation.
5.5.2.2.4. Choosing Kernels¶
The key sensitivity calculation inputs are kernels. We need two kernels:
a kernel to quantify the dis/similarity between parameter values;
a kernel to classify the loss function values.
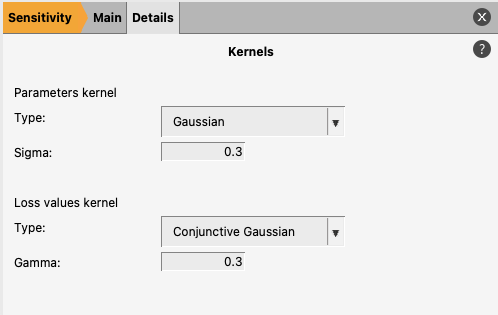
The default kernels should be sufficient in most scenarios.
The default parameter values kernel applies a Gaussian kernel to the parameter values. This measures the distance between parameter values and applies no underlying assumption on the shape of the distribution of parameter values.
The default loss values kernel is a conjunctive-Gaussian type kernel which places the bulk of its distribution on the lowest loss values. This helps identify small local sensitivities near the minima we are interested in which might otherwise be drowned out by large global sensitivities.
5.5.3. Sensitivity results¶
Save and run the calculation:
The calculation should take a couple of seconds. You can see progress and get an estimated completion time from the progress bar printed in the logfile.
5.5.3.1. Graphical results¶
Once complete the GUI will show 2 summary plots:
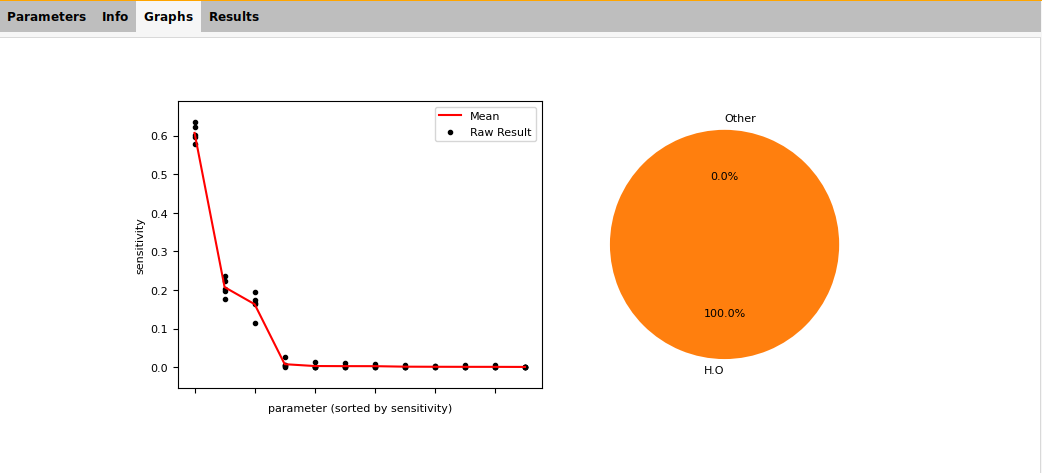
Fig. 5.10 Example sensitivity result summary provided by the GUI¶
The scatter plot shows:
the sensitivity result for each parameter for each repeat (black dots);
the mean sensitivity for each parameter (red line).
This does not identify particular parameters, but one can see:
the number of parameters which dominate the sensitivity;
if there is a large spread between repeats (meaning more samples are needed).
In this example:
we detect significant sensitivity for only three parameters;
the spread between repeats is robust and does not influence the ordering of parameters.
The pie chart shows the allocation of sensitivity to different groups. For ReaxFF, the groups are combinations of atoms and blocks. Here we only analyzed parameters in the H.O bond block, so all of the sensitivity is in that group.
5.5.3.2. Sensitivity values¶
 panel if it is open
panel if it is openTip
You might have to resize your window to see the Sensitivity and Description columns
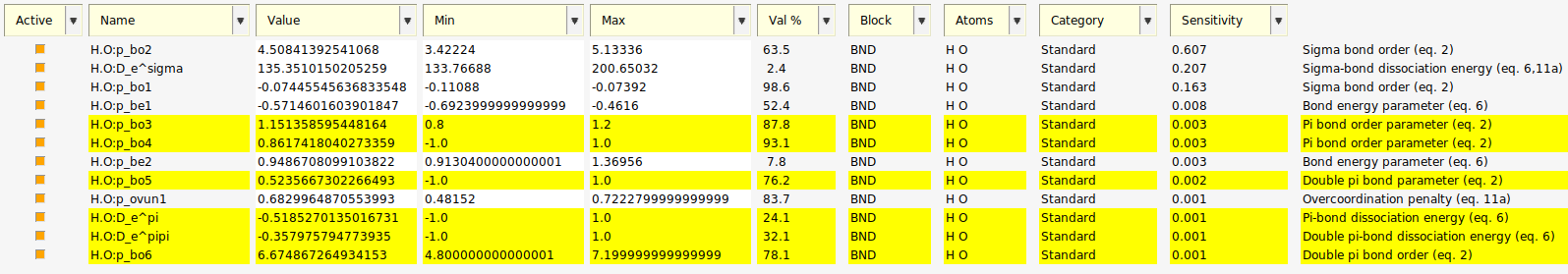
Fig. 5.11 12 active parameters and their calculated sensitivities with parameters related to π-bonds selected.¶
Each active parameter has been given the average sensitivity value from the repeats (seen as the red line on the plot).
In our results we see:
negligible sensitivity was detected for the π-bond related parameters;
H.O:p_be1, H.O:p_be2 and H.O:p_ovun1 are also found to be insensitive;
Most of the sensitivity is divided between three σ-bond related parameters.
5.5.3.3. Results files¶
The results for a sensitivity calculation have the following structure:
jobname.results
├── sampling_results (if samples generated)
│ ├── settings_and_initial_data
│ └── optimization (directory to chose when loading samples)
├── settings_and_initial_data
│ └── data_sets
└── sensitivity
├── calculation_result.npz
├── parameter_interface_with_sensitivities.yaml
├── sensitivity_per_parameter.png
├── sensitivity_summary.png
└── sensitivity_summary.txt
calculation_result.npz contains the calculation results and the inputs which can be loaded in Python:
from scm.glompo.analysis.hsic import HSICResult if __name__ == '__main__': result = HSICResult.load('path/to/calculation_result.npz')
For more details see the API documentation
parameter_interface_with_sensitivities.yaml is the normal parameter interface YAML file with a
sensitivitymetadata key for all the parameters. The key is blank if the parameter was not ‘active’ for the calculation.--- name: O.O:p_ij^xel1 value: 0.0 range: - -1.0 - 1.0 is_active: false atoms: - O - O block: BND block_index: 15 category: Expert description: eReaxFF param for adjusting number of electrons available to host atom equation: '27' sensitivity: '' --- name: H.O:D_e^sigma value: 135.35101502052595 range: - 133.76688000000001 - 200.65031999999997 is_active: true atoms: - H - O block: BND block_index: 0 category: Standard description: Sigma-bond dissociation energy equation: 6,11a sensitivity: 0.2074254860483457 ---
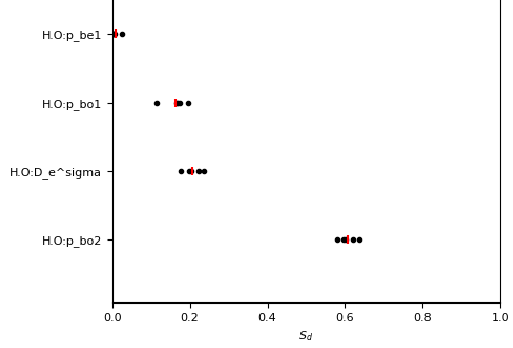
sensitivity_per_parameter.png is a visual summary of sensitivity values for every parameter and every repeat.
sensitivity_summary.png (and gui_sensitivity_summary.png) are discussed in Graphical results.
sensitivity_summary.txt is a textual summary of the calculation inputs and sensitivity values for every parameter.
HSIC Result ================================================== Calculation Description: TrainingSet -------------------------------------------------- No. Factors 12 No. Samples 500 No. Bootstraps 5 Sample with replacement True Includes reweight calc. No -------------------------------------------------- Inputs Kernel GaussianKernel sigma 0.3 Outputs Kernel ConjunctiveGaussianKernel gamma 0.3 -------------------------------------------------- Factor Rankings: 1. H.O:p_bo2 0.607±0.020 2. H.O:D_e^sigma 0.207±0.021 3. H.O:p_bo1 0.163±0.026 4. H.O:p_be1 0.008±0.009 5. H.O:p_bo3 0.003±0.005 6. H.O:p_bo4 0.003±0.005 7. H.O:p_be2 0.003±0.003 8. H.O:p_bo5 0.002±0.003 9. H.O:p_ovun1 0.001±0.001 10. H.O:D_e^pi 0.001±0.002 11. H.O:D_e^pipi 0.001±0.002 12. H.O:p_bo6 0.001±0.001 ==================================================
5.5.3.4. Using the results for optimization¶
The purpose of sensitivity analysis is to guide parameter selection for optimization.
Our results suggest that only three parameters have an effect on the loss function.
We have run the optimization version of this tutorial 10 times:
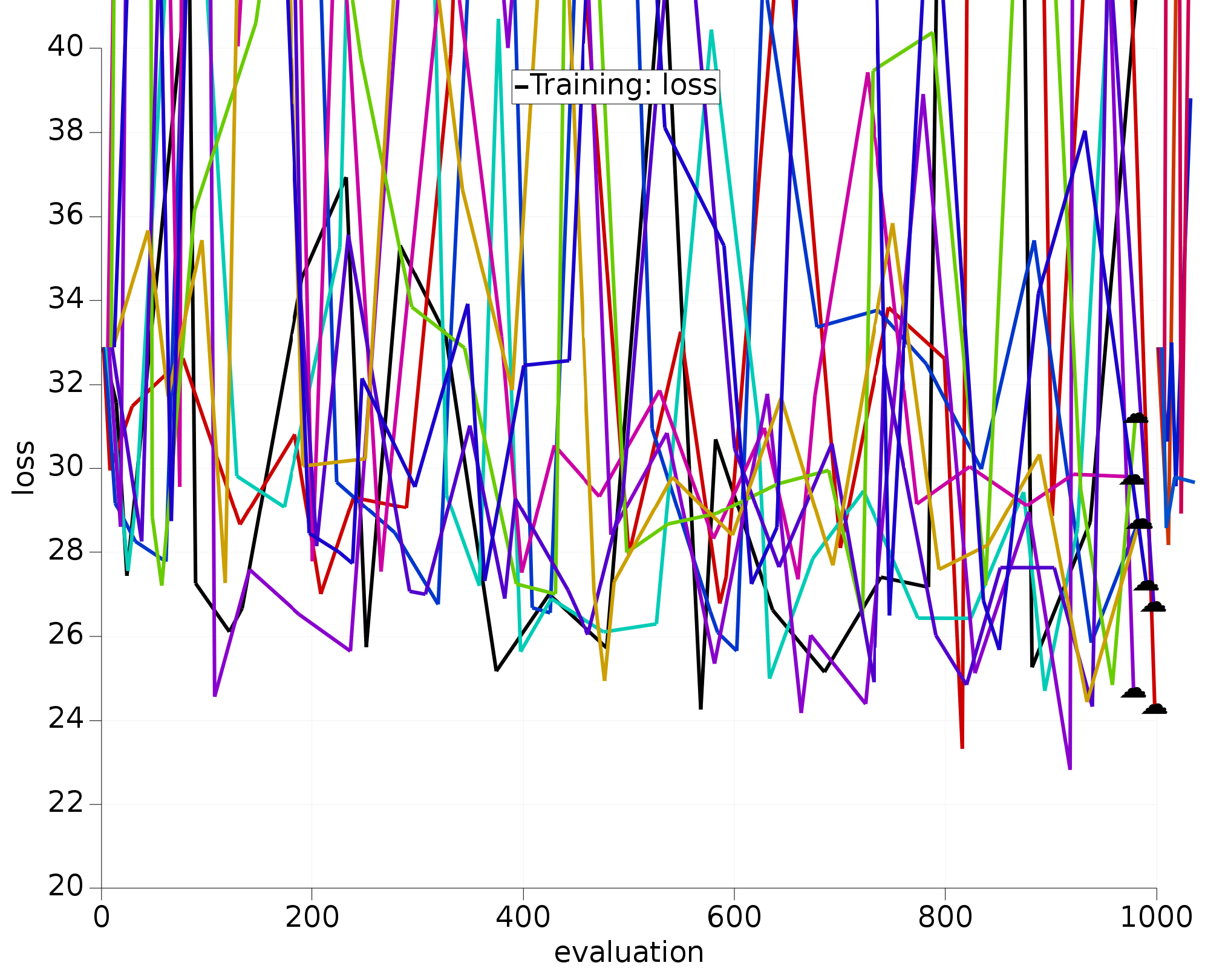
Fig. 5.12 10 optimizations using all 12 H.O parameters.¶
We have also run the optimization with the exact same settings, but with only the three most sensitive parameters active; also repeated 10 times:
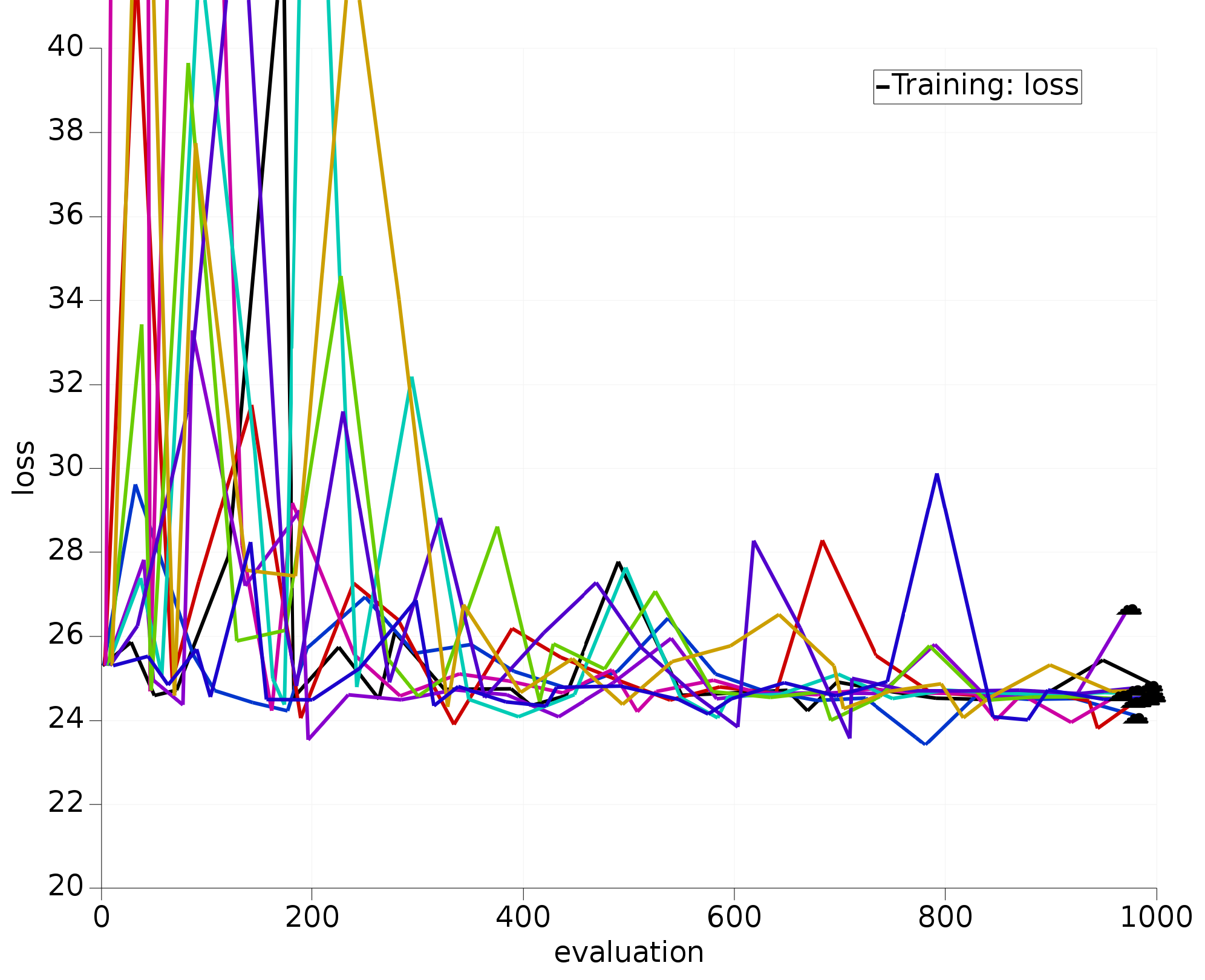
Fig. 5.13 10 optimizations using 3 most sensitive H.O parameters.¶
The optimizers working only with the most sensitive parameters clearly:
converge much faster;
all find similar/better loss function values than the 12D optimizers.
See also
To learn about running multiple optimizers at once see Parallel optimizers.
5.5.4. Limits of the sensitivity calculation¶
There are several factors which affect the accuracy of the sensitivity calculation:
having a sufficient number of samples is the most important factor, particularly in high-dimension;
more repeats will provide a more accurate sensitivity average;
make subsets large enough;
kernel hyperparameters should not be overly restrictive;
It is also important to be aware of the conditions which invalidate a sensitivity calculation. If any of the following changes are made after a sensitivity calculation, then you should no longer rely on the sensitivity values:
adding/removing/editing items in the training set;
changing sigma/weight values in the training set;
changing job collection settings;
changing a parameter to in/active;
changing a parameter range.
Warning
If a modification is small, one might reasonably expect the sensitivity values to remain reasonably unchanged, however, there is no guarantee that this is true. The sensitivity profile might be radically different.
Finally, there is a trade-off when choosing the number of parameters to optimize. When activating fewer parameters the optimization becomes easier and faster, but, this usually comes at the cost of making the minima worse. This is because there is less flexibility in the potential function to fit itself to all the training data. Worse minima is not always a bad thing. Using fewer parameters can reduce the problem of overfitting.
5.5.5. Some rules of thumb¶
When picking parameters be guided by sensitivity, but consider including several less sensitive parameters in your selection (if their selection makes intuitive sense). For example, in a training set with no π-bonds, do not activate π-bond related parameters but a low sensitivity σ-bond parameter may make sense.
Roughly, for a training set with \(n\) items, do not fit more than \(\sqrt{n}\) parameters.
For a successful production-quality reparameterization you may need to optimize in several steps: first with very sensitive parameters and then only with moderately sensitive ones.
If sensitivities are very imbalanced (a handful of parameters are much more sensitive then the others) consider optimizing in multiple steps or redesigning/reweighing the training set to balance the sensitivities better and make the loss function easier for the optimizers to minimize.
5.5.6. Sensitivity job in Python¶
In the examples directory, the file sensitivity.py shown below illustrates how to
run the sensitivity calculation
create a new parameter interface with only the 3 most sensitive parameters active
run an optimization job using the new parameter interface
The code below uses the ParAMSResults.get_ordered_parameters() function
to extract a list of parameter names sorted by sensitivity. For more possibilities, see the ParAMSResults API.
#!/usr/bin/env amspython
from scm.plams import *
from scm.params import *
import os
def main():
init() # initialize PLAMS workdir
examples_dir = os.path.expandvars("$AMSHOME/scripting/scm/params/examples/ReaxFF_water")
sensitivity_job = ParAMSJob.from_inputfile(examples_dir + "/sensitivity.in", name="sensitivity")
sensitivity_job.run()
# or load a finished sensitivty job using ParAMSJob.load_external:
# sensitivity_job = ParAMSJob.load_external('plams_workdir/sensitivity/results')
# create a new interface with top 3 sensitive parameters activated
new_interface_path = "top3_activated_interface.yaml"
create_interface_with_most_sensitive_parameters(sensitivity_job, new_interface_path, N=3)
# load the optimization job from examples directory but use the new parameter interface
optimization_job = ParAMSJob.from_inputfile(examples_dir + "/params.in", name="optimization")
optimization_job.parameter_interface = new_interface_path
optimization_job.run()
finish()
def create_interface_with_most_sensitive_parameters(job: ParAMSJob, output_fname: str, N=3):
"""
Gets the N most sensitive parameters from the previous sensitivity job,
stores resulting interface in output_fname
"""
top_n_sensitive_parameters = job.results.get_ordered_parameters()[:N]
interf = job.results.get_initial_parameter_interface()
for p in interf:
p.is_active = p.name in top_n_sensitive_parameters
interf.yaml_store(output_fname)
if __name__ == "__main__":
main()