Nudged Elastic Band (NEB)¶
In this tutorial we will use the climbing-image Nudged Elastic Band method (NEB) to identify the minimum energy path of:
Transition state of a chemical reaction¶
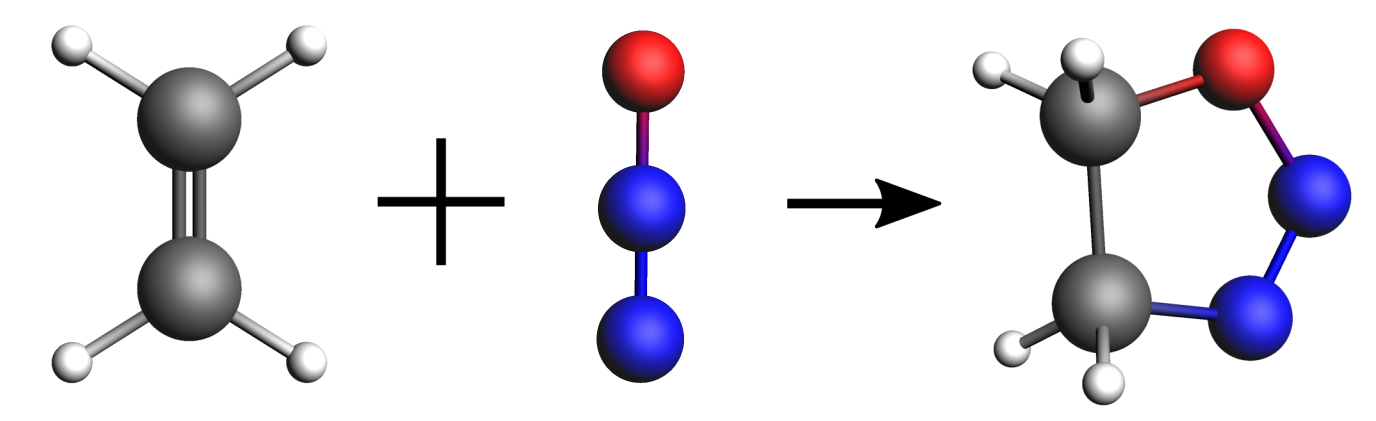
In this first example, we will find the minimum energy reaction path and transition state of the following reaction:
We will be using the DFTB engine with the GFN1-xTB model. This is a computationally fast method (which is convenient for the purposes of a tutorial) but it is not very accurate in predicting reaction energies. For better accuracy, using the DFT engine ADF (or BAND) is recommended.
Setting up and running the calculation¶
You can set up and run the calculations for this tutorial using either the Graphical User Interface (GUI), the python library PLAMS or a bash script.
This will open a new AMSinput window.
 →
→ 
We will use the GFN1-xTB model, which is the default model as of the AMS2020 release. Your AMSinput window should look like this:
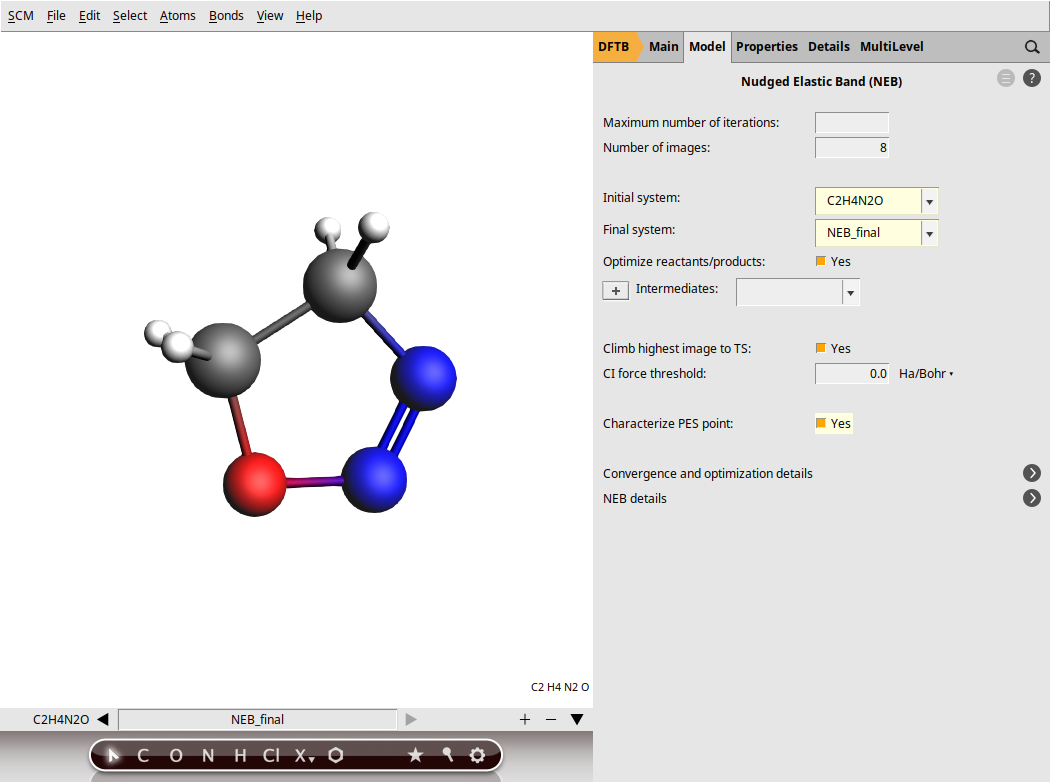
When setting up an NEB calculation we need to specify two systems: the initial and final states of the reaction. The NEB algorithm will then generate a set of images by interpolating between the initial and final systems. This will be the initial approximation of the reaction path, which will be optimized during the NEB calculation.
NEB_initial.xyz and NEB_final.xyzYou can easily switch between molecule tabs using the arrows at the bottom of the molecule drawing area. Here, you’ll also see the molecule names, such as C2H4N2O for the first molecule and NEB_final for the second. Note that the GUI has the following naming convention:
Main System: The first molecule is considered the main system, and is often automatically assigned its chemical formula as the name.
Additional Molecules: For any additional molecules, the GUI uses by default the name from their original XYZ files.
Important
In NEB calculations, the order of the atoms in the initial and final system should be the same (if you provide an intermediate system, you should use a consistent atom-ordering for that too). The order of the atoms should be consistent because the images-interpolation algorithm maps the n-th atom of the initial system to the n-th atom of the final system.
You can see the indices of the atoms by clicking in the menu bar on View → Atom Info → Name → Show. It is possible to change the order of the atoms in the Coordinates panel (in the panel bar: Model → Coordinates) using the Move atom(s) option.
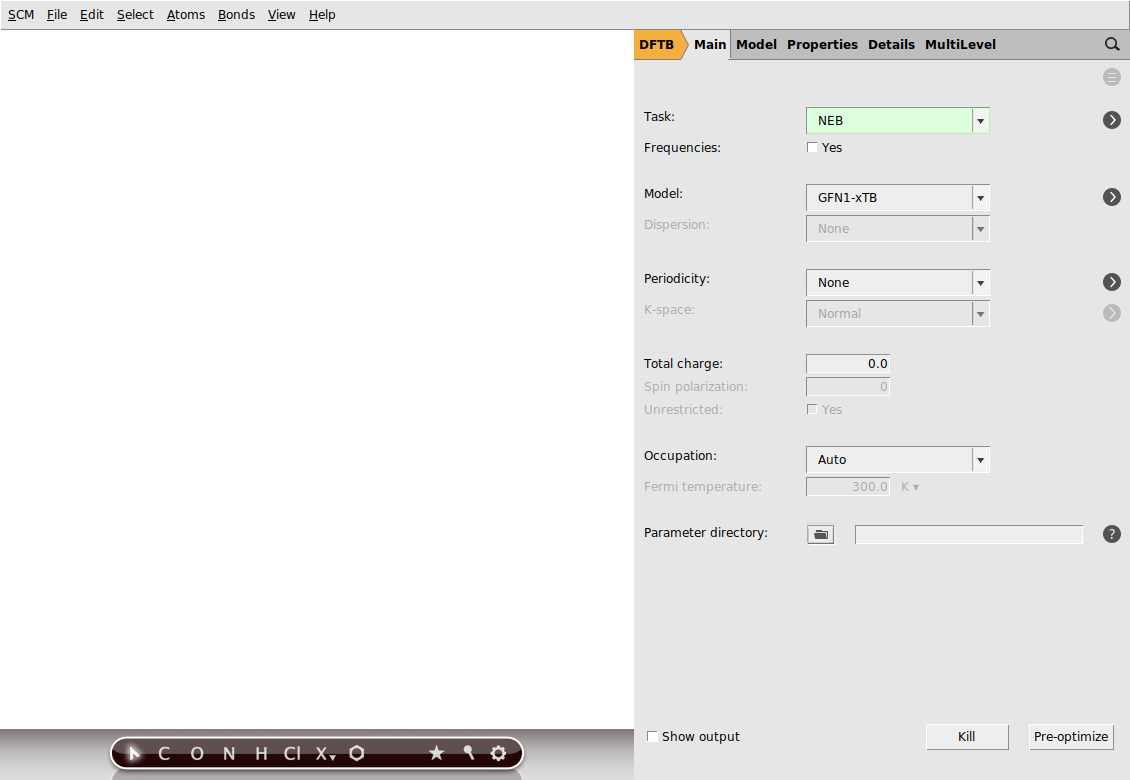
Now, go to the NEB details panel where we will set up the NEB calculation:
 next to Task → NEB to go to the NEB details panel
next to Task → NEB to go to the NEB details panelYour AMSinput window should look like this:
Tip
From most AMSinput panels you can jump to the relevant section of the user manual by clicking on  , which is located in the top-right corner of the panel.
, which is located in the top-right corner of the panel.
We are now ready to run the calculation:
In the logfile you can monitor the progress of your NEB calculation:
A NEB calculation consists of several steps, which are automatically executed one after the other:
first, the two end points (the initial and final molecules) are optimized (in the logfile, look for
Optimizing initial stateandOptimizing final state)then the NEB reaction path will be optimized (in the logfile, look for
NEB Path Optimization). During the reaction path optimization, the highest-energy image on the path will climb to the transition statefinally, a single point calculation for the TS is performed (in the logfile:
Final calculation for highest-energy image). If the option Characterize PES point is on, then the lowest-lying normal modes will be calculated in order to validate the TS (the TS should have exactly one imaginary frequency). Some information on the reaction path is printed at the end of the logfile:NEB found a transition state! TS barrier height from the left 0.02576078 Hartree 16.165 kcal/mol 67.635 kJ/mol TS barrier height from the right 0.08632064 Hartree 54.167 kcal/mol 226.635 kJ/mol
The following bash script performs an NEB calculation using the AMS driver and the DFTB engine. The input options for the AMS driver described in the AMS driver manual, while the DFTB manual describes the input options for the DFTB Engine block.
#!/bin/sh
"$AMSBIN/ams" << eor
Task NEB
Properties
PESPointCharacter Yes
End
# The initial system (no header)
System
Atoms
N 1.88630912 -0.34204867 -1.59424245
N 1.14203025 -1.15084766 -1.95458206
O 0.07639739 -1.27682918 -1.40578886
C 0.74766633 1.24132374 1.29097263
C -0.52735637 0.87051548 1.20080269
H 1.08791192 2.15092371 0.80682956
H -1.23438622 1.47569873 0.64278673
H -0.86675778 -0.04061999 1.68265930
H 1.45534611 0.63457332 1.84647164
End
End
# The final system should be called 'final'
System final
Atoms
N 1.44280525 0.39326405 0.02115802
N 0.56588608 -0.32983790 -0.53167497
O -0.68785300 -0.26603404 -0.00725496
C 0.86550207 1.12517445 1.10513146
C -0.57889386 0.65549932 1.05974328
H 0.94852175 2.21696742 0.91684905
H -1.26384434 1.51200042 0.88137995
H -0.85746564 0.15513847 2.01191277
H 1.35845740 0.84559176 2.06071421
End
End
Engine DFTB
Model GFN1-xTB
EndEngine
eor
Important
In NEB calculations, the order of the atoms in the initial and final system should be the same (if you provide an intermediate system, you should use a consistent atom-ordering for that too). The order of the atoms should be consistent because the images-interpolation algorithm maps the n-th atom of the initial system to the n-th atom of the final system.
To run the calculation:
NEB.runchmod +x NEB.run./NEB.run > outIn the following python script (using the PLAMS library) we set up a NEB calculation, run it, and extract some results from the binary results files.
# Load the molecules from file
initial_mol = Molecule("NEB_initial.xyz")
final_mol = Molecule("NEB_final.xyz")
settings = Settings()
# Input options for the AMS driver
settings.input.ams.Task = "NEB"
settings.input.ams.Properties.PESPointCharacter = "Yes"
# Input options for the engine (DFTB in this case)
settings.input.DFTB.Model = "GFN1-xTB"
# You can pass multiple systems to an AMSJob by using a dictionary of molecules.
# The key of the dictionary will be used as the header of the 'System' block
molecules = {"": initial_mol, "final": final_mol}
# Create and run the job
job = AMSJob(settings=settings, molecule=molecules, name="NEB")
results = job.run()
if job.ok():
print("Successful NEB calculation!")
# Collect the results
pes_point_character = results.readrkf("AMSResults", "PESPointCharacter", file="NEB_TS_final")
molecule_ts = results.get_main_molecule()
# Results on the binary files are in atomic units
left_barrier = results.readrkf("NEB", "LeftBarrier")
right_barrier = results.readrkf("NEB", "RightBarrier")
# Convert units using the Units class
left_barrier_kcalmol = Units.convert(left_barrier, "au", "kcal/mol")
right_barrier_kcalmol = Units.convert(right_barrier, "au", "kcal/mol")
# Print the results
print(f"PES Point character: {pes_point_character}")
print("Geometry of the TS:")
print(molecule_ts)
print(f"Left TS barrier : {left_barrier_kcalmol:.6f} [kcal/mol]")
print(f"Right TS barrier: {right_barrier_kcalmol:.6f} [kcal/mol]")
else:
print("NEB calculation not successful...")
The options for the AMS driver and for the DFTB engine are specified in the Settings object object. The various input keys are described in the AMS driver manual and DFTB manual. See the AMS interface section of the PLAMS manual for more details.
Important
In NEB calculations, the order of the atoms in the initial and final system should be the same (if you provide an intermediate system, you should use a consistent atom-ordering for that too). The order of the atoms should be consistent because the images-interpolation algorithm maps the n-th atom of the initial system to the n-th atom of the final system.
To run the PLAMS script:
NEB_initial.xyz and NEB_final.xyzNEB.py$AMSBIN/plams NEB.pyTo improve the initial approximation of the reaction path in an NEB calculation, you can (optionally) provide an intermediate system.
Another important NEB option is the number of images. In case of problematic NEB path optimization convergence, using more images might help (note that the computation time increases with the number of images used).
You can read more about the various NEB options in the AMS manual.
Results of the calculation¶
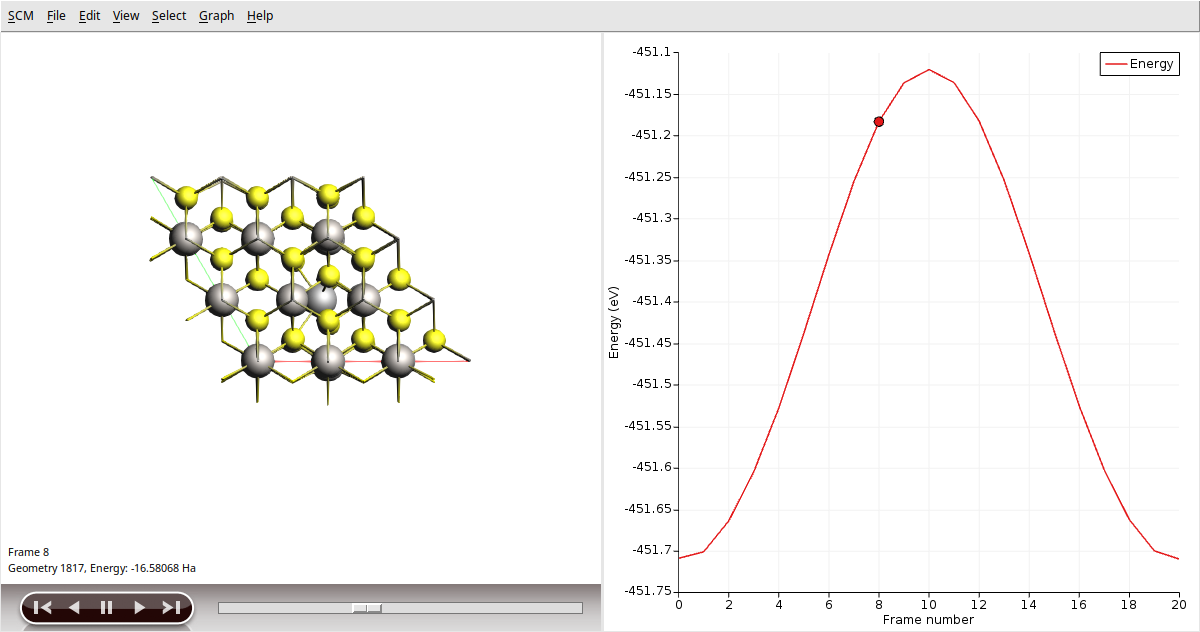
Now, let’s visualize the reaction path computed by NEB:
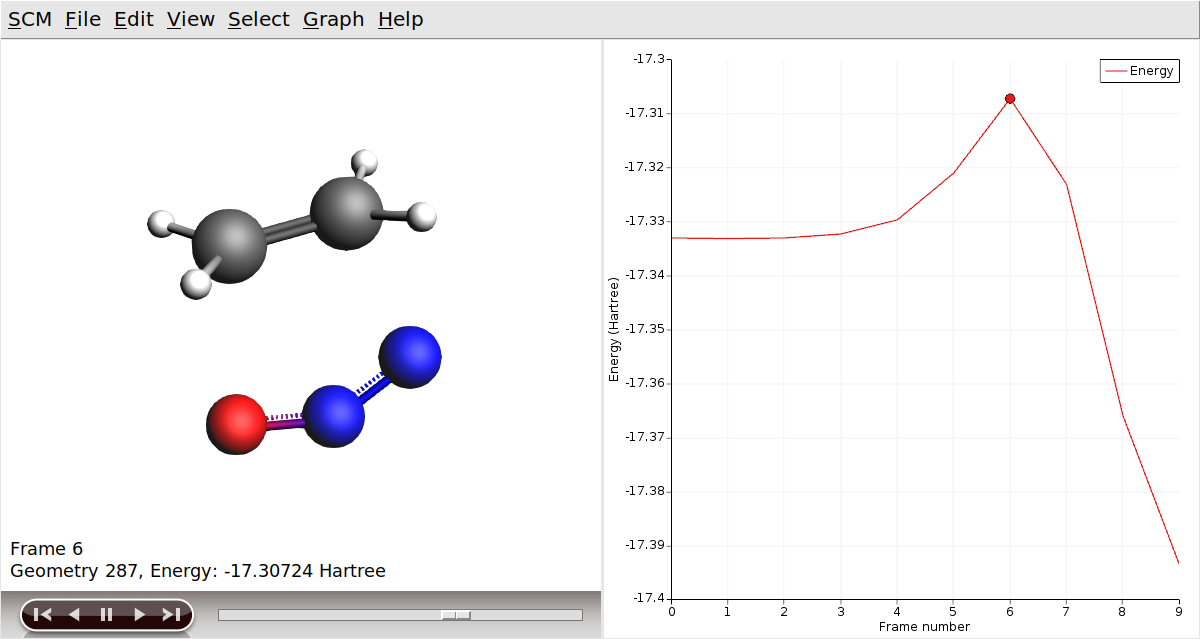
In AMSmovie, you can click on play (or drag the slide-bar) so see the reaction happening. On right-hand side, the energy and gradients of the images in the NEB reaction path are plotted.
The transition state is characterized by having one imaginary frequency. Let’s visualize the normal modes of the transition state geometry with AMSspectra:
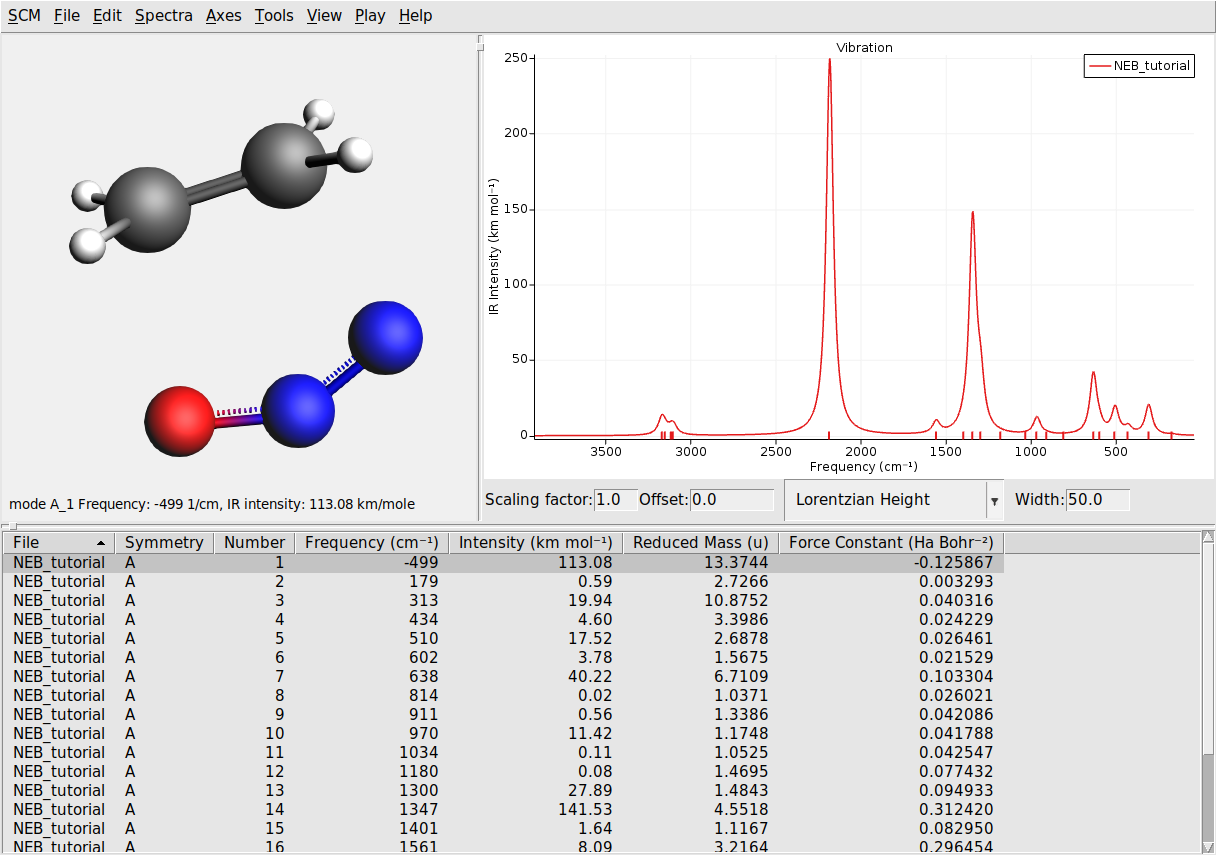
Here you will see the computed normal modes for the TS geometry. Notice that there is one negative frequency (imaginary frequency are shown as negative numbers).
The corresponding normal mode will be displayed in the molecule-visualization area. This normal mode gives you an idea of how the atoms are moving as they cross the transition state.
In the folder where you executed your script you will find a file out, which contains the text-output of the calculation, and a folder called ams.results containing binary results of the calculation.
At the end of the output file out you will find a section summarizing the results of the NEB calculation:
NEB found a transition state!
------------------------------------------------------------
TS barrier height from the left 0.02581511 Hartree
16.199 kcal/mol
67.778 kJ/mol
TS barrier height from the right 0.08637041 Hartree
54.198 kcal/mol
226.765 kJ/mol
Reaction energy -0.06055530 Hartree
-37.999 kcal/mol
-158.988 kJ/mol
------------------------------------------------------------
Transition state geometry
--------
Geometry
--------
Atoms
Index Symbol x (angstrom) y (angstrom) z (angstrom)
1 N 1.76794468 0.02199401 -0.76530642
2 N 0.86568320 -0.56021023 -1.14090273
3 O -0.28459962 -0.76023024 -0.92163051
4 C 0.89983351 0.97770099 0.84893165
5 C -0.38912967 0.55813520 0.80607959
6 H 1.18303105 1.94163319 0.44757192
7 H -1.15509240 1.14579400 0.31965225
8 H -0.73127815 -0.27639812 1.40152984
9 H 1.61076814 0.51427067 1.51998360
The folder ams.results contains:
a text file called
ams.logcontaining a very concise summary of the calculation’s progress during the run.the main binary results file
ams.rkf, containing the reaction path computed in the NEB calculation.the engine results file
NEB_TS_final.rkfcorresponding a single point calculation at the transition state geometry. It contains, among other properties, the normal modes.
You can explore the content of the rkf binary results files using the kfbrowser utility.
$AMSBIN/kfbrowser ams.results/NEB_TS_final.rkfThe binary results of the calculation can also be visualized with the GUI modules:
$AMSBIN/amsmovie ams.results/ams.rkf$AMSBIN/amsspectra ams.results/NEB_TS_final.rkfThis is the output printed by the PLAMS script:
[11:48:45] PLAMS working folder: [...]
[11:48:45] JOB NEB STARTED
[11:48:45] JOB NEB RUNNING
[11:48:47] JOB NEB FINISHED
[11:48:47] JOB NEB SUCCESSFUL
Successful NEB calculation!
PES Point character: transition state
Geometry of the TS:
Atoms:
1 N 1.762731 0.038376 -0.779381
2 N 0.866215 -0.561703 -1.142377
3 O -0.278098 -0.778480 -0.906970
4 C 0.901016 0.975210 0.848982
5 C -0.389988 0.560115 0.803367
6 H 1.186939 1.941923 0.456712
7 H -1.151130 1.151965 0.314116
8 H -0.738634 -0.269125 1.402220
9 H 1.608109 0.504408 1.519239
Left TS barrier : 16.199224 [kcal/mol]
Right TS barrier: 54.198250 [kcal/mol]
[11:48:47] PLAMS run finished. Goodbye
In the folder where you executed your script you will find a newly created folder plams_workdir containing the results of the calculations. The folder plams_workdir/NEB contains the results of the job NEB. Inside this folder you will find all the files generated by AMS, including the binary results files ams.rkf and NEB_TS_final.rkf.
The binary results of the calculation can also be visualized with the GUI modules:
$AMSBIN/amsmovie plams_workdir/NEB/ams.rkf$AMSBIN/amsspectra plams_workdir/NEB/NEB_TS_final.rkfIonic diffusion in a solid¶
In this second example, we will determine the minimum energy path and the activation energy of Li diffusion in a typical cathode materials LiTiS2.
We will use the ML potential engine with the M3GNet universal machine learning potential. Although M3GNet was trained to reproduce ground state structures of the Materials Project database, it can also be accurate to describe transition states, as we will demonstrate in this tutorial. This tutorial can be reproduced with the periodic DFT engine BAND.
First, make sure to install the M3GNet package inside AMS as described in this tutorial.
Layer LiTiS2 cathode material¶
In some Li-based cathode materials, the diffusion of Li ions is tightly connected to the content of Li in the materials. We would like to demonstrate this by computing the diffusion barrier of Li in LiTiS2 with 1 and 2 Li vacancies. In layer LiTiS2, the lithium ions are octahedrally coordinated between the S−Ti−S sandwich layers.
Li diffusion in LiTiS2¶
To perform the NEB calculation we will use the graphical user interface and perform a 3-step calculation: initial state, final state, and NEB.
Step 1: Initial state¶
Download the crystal structure of layer LiTiS2.
This will open a new AMSinput window. Let’s perform a geometry and cell optimization of the initial structure.
 →
→ 
 next to Task → Geometry Optimization
next to Task → Geometry OptimizationNote
When using M3GNet it is highly recommended to include dispersion via the addon option. In the following, we will build the final state, and the NEB calculation from the initial state calculation and therefore the D3 dispersion will be included throughout.
Save the job (File → Save As..), give it a name (LiTiS2_relax), and run the job Job → Run. This should take a few minutes. When asked to update coordinates with bonds and data say yes.
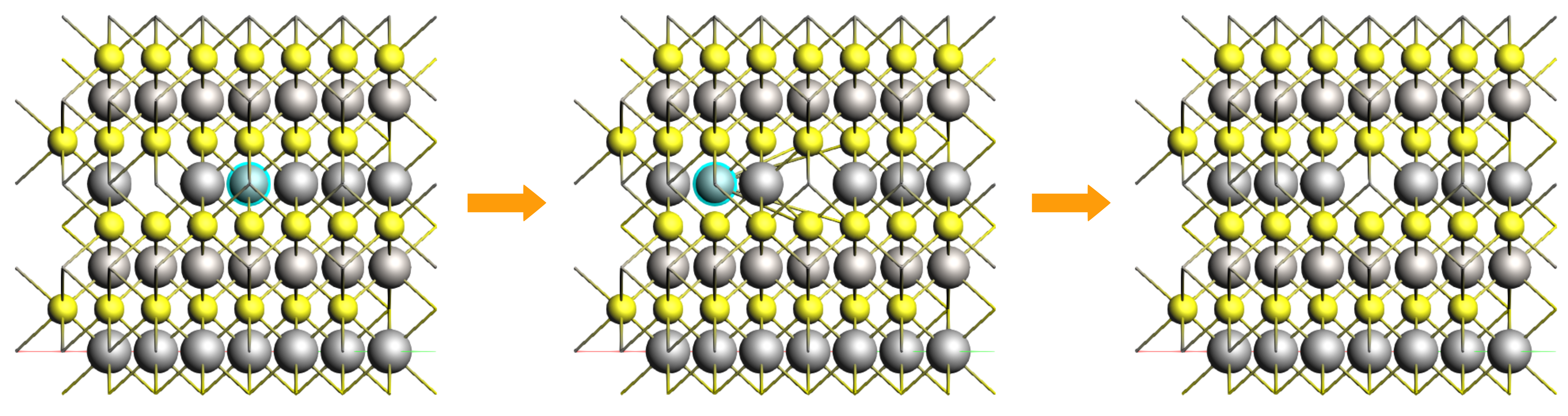
Orient the crystal along the x-axis (Ctrl+1), then hold the right mouse button and slightly move the molecule for a better view of the middle Li atom in the top layer (index 25). Select this Li atom and press the ‘Delete’ key to remove it.
 next to Task → Geometry Optimization
next to Task → Geometry OptimizationSave the job (File → Save As..), give it a name (LiTiS2_init), and run the job Job → Run. When asked to update coordinates with bonds and data say yes.
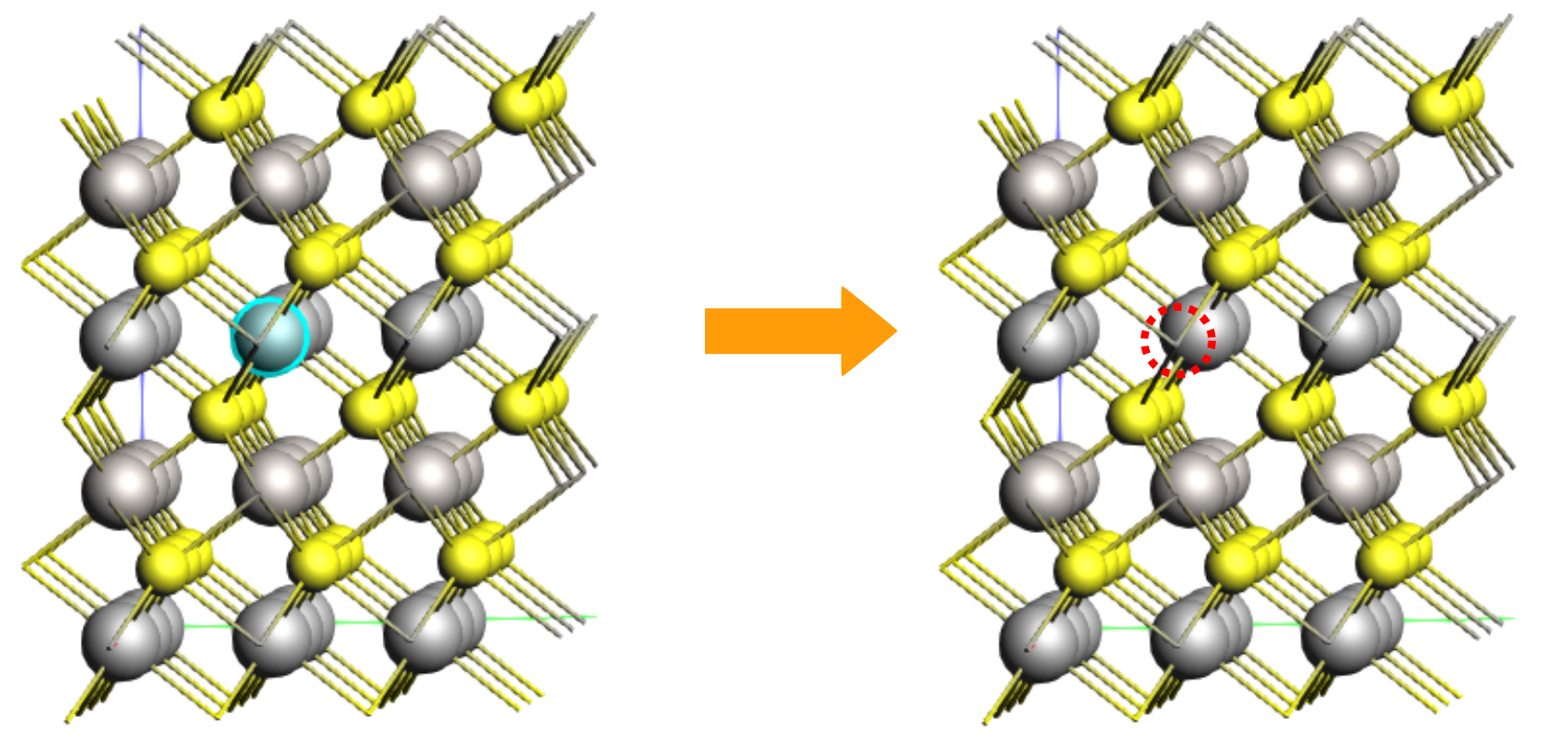
Let’s use this structure as the starting point for the final calculation. Save the new job (File → Save As..) with the name LiTiS2_final.
Step 2: Final state¶
Note
When you perform NEB in a crystal it is recommended to keep the cell dimension between the initial and final states.
Save and run the calculation without changing the settings to perform a geometry optimization of the final state.
When asked to update coordinates with bonds and data say yes.
Check that the energy of the initial and final state are almost identical using SCM → Output (section CALCULATION RESULTS).
You should values close to -16.59999540 hartree for the initial state and -16.60003303 hartree for the final state.
Step 3: NEB¶
We now have the initial and finial state geometries and we can proceed to the NEB calculation. We will modify the initial state.
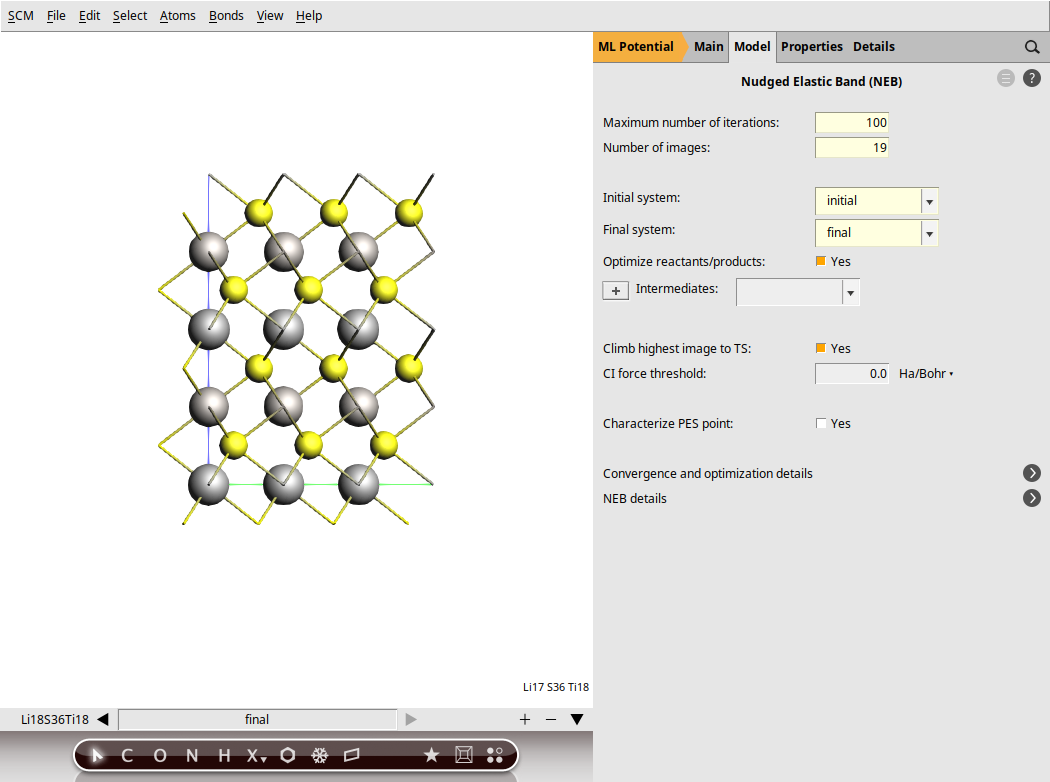
We can now define the settings for the NEB calculation.
 next to Task → NEB
next to Task → NEBNotice that the molecule area has been cleared. What happened is that AMS created a new empty molecule, named Mol-1 by defaults. You can switch between the initial state Li18S36Ti18 and the final state Mol-1 by clicking the small black triangles at the bottom of the molecule area. There are several ways to define the final system. We will copy and paste the final system we have previously prepared. Keep the LiTiS2_NEB input window opened, with the empty molecule area Mol-1.
Note
Alternatively, you can save the coordinates of the final state and load it to Mol-1.
The NEB is now ready. Save the calculation. This will automatically rename the molecules as initial and final.
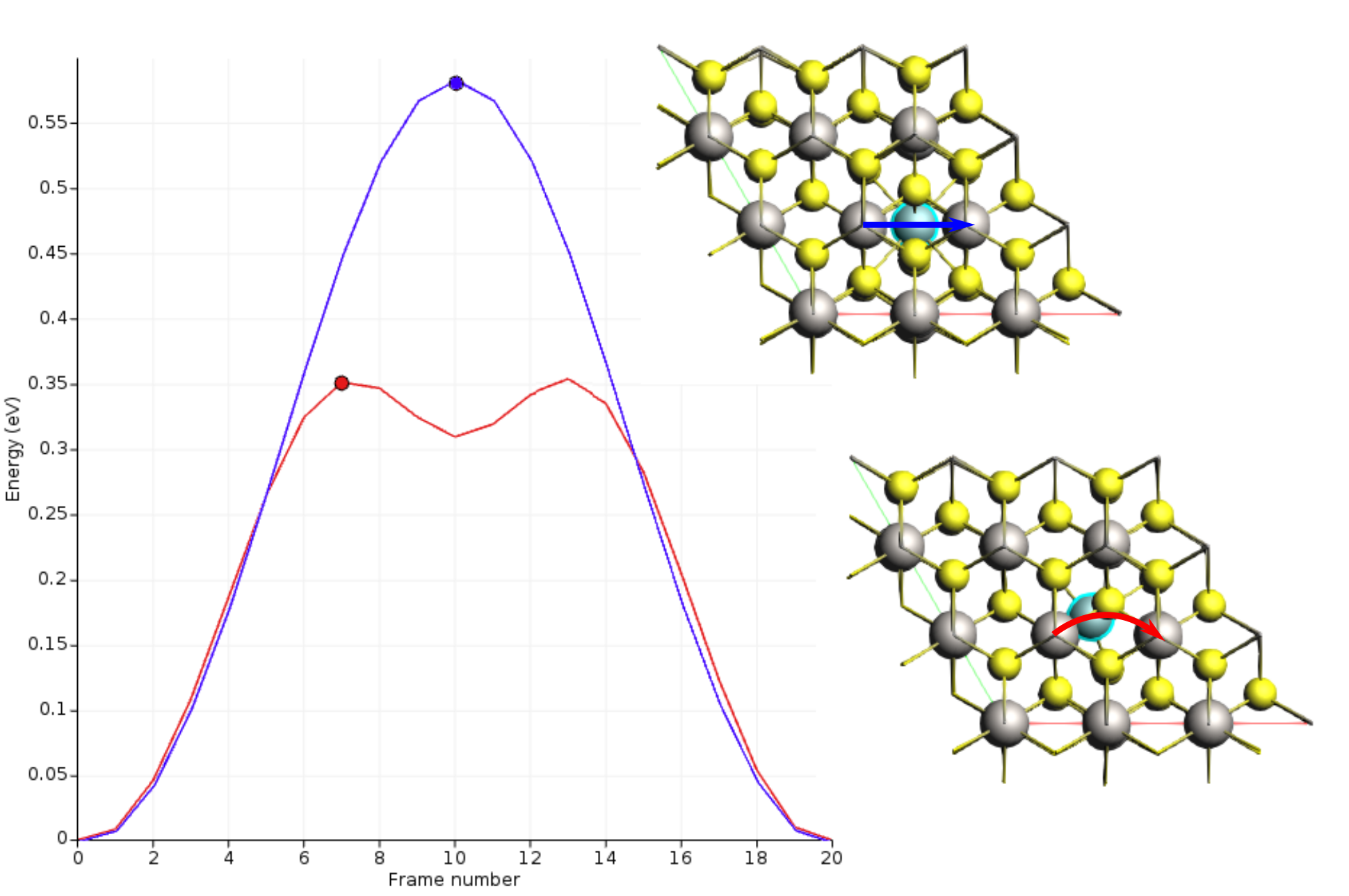
Run the calculation. This might take a few more minutes as AMS will perform a simultaneous optimization of all the 17 images by minimizing the forces parallel to the reaction path. Once the calculation is finished, select LiTiS2_NEB in AMSjobs and click SCM → Movie. You can re-orient the crystal and use the slidebar under the molecule area to appreciate the diffusion along the minimum energy path. If you double click the y-axis label of the graph, you can switch to your prefered units. We found the activation energy EA = 0.59 eV.
For a streamlined approach, you can automate the entire workflow using the following Python script: here.
Li diffusion in LixTiS2¶
To explore the effect of lithium concentration on diffusion, begin with the structure from step 3. Remove a neighboring lithium atom (index 9) and at the end increase the NEB iterations to 200. If you’re using the Python script, update it to reflect these changes. This change should reduce the activation energy by approximately a factor of 2. Interestingly, while the lithium migration trajectory remains linear when all neighboring lithium sites are occupied, a vacant neighbor site causes the path to curve, passing through a tetrahedral site and creating a shallow energy minimum.