QTAIM (Bader), localized orbitals and conceptual DFT¶
QTAIM analysis of an Adenine–Thymine base pair¶
- Start ADFinput
Next we need the structure of the Adenine–Thymine (AT) base pair:
- Click on the Structure toolSelect DNA → ATClick on the molecule drawing area to add the AT base pair

You should now see the AT base pair in the molecule drawing area.

Now we want to activate the QTAIM analysis to find the critical points and bond paths. To find where this option is located, search for it:
ADFinput will switch to the panel that displays the option you are looking for (to calculate the AIM critical points and paths). The matching input options will be marked with blue italic text. Note that the input option search will restrict the search to panels that belong to the current method (ADF, BAND, DFTB, …)
- Tick the Perform QTAIM analysis check-boxSet the Analysis level to Full (to get atomic properties, basins and other descriptors in addition to the critical points and paths)

Now we are ready to run the calculation.
- To run the calculation: File → Run
A dialog will pop up in which you must specify a filename to use for your job, for example ‘AT’:
- Enter ‘AT’ as a Filename, press the Save button
After hitting the save button the calculation will start. You will get a new ADFjobs window that allows you to manage your jobs and keep track of their state (for example, queued or running). For a running job, there will be two lines showing the progress. To get more information you can show the logfile in a new window:
- Click on the two lines showing the progress
Depending on your computer, the calculation should be ready after a few minutes at most.
Once the calculation is finished, use ADFview to visualize the results. To visualize the critical points and bond paths:
- In ADFView, click on Properties → QTAIM (Topology)
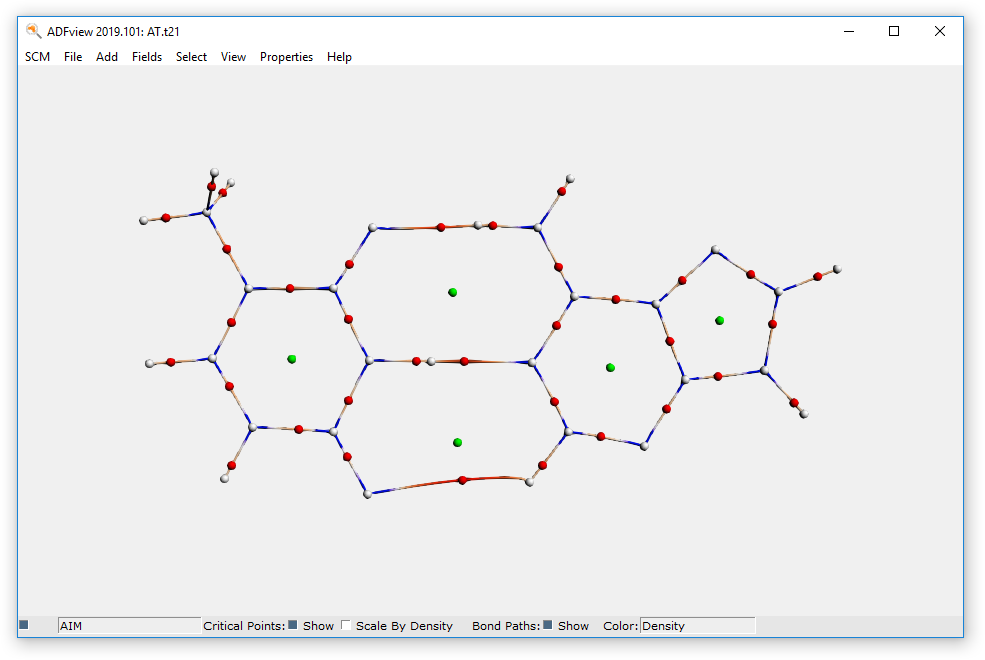
The critical points and bond paths are shown (the molecule balls and sticks representation is hidden). The different types of critical points (atom CP, bond CP, ring CP and cage CP) are indicated by different colors. The bond paths are colored by density, by default. You can get extra information about a CP or a point along the bond path by clicking on it.
We will now add a cut-plane showing the electronic density:
- Click on Fields → Grid → MediumClick on Add → Cut Plane: ContoursIn the bar at the bottom of ADFView, click on Select Field → Density → Density SCFChange the number of contour lines from 15 to 30
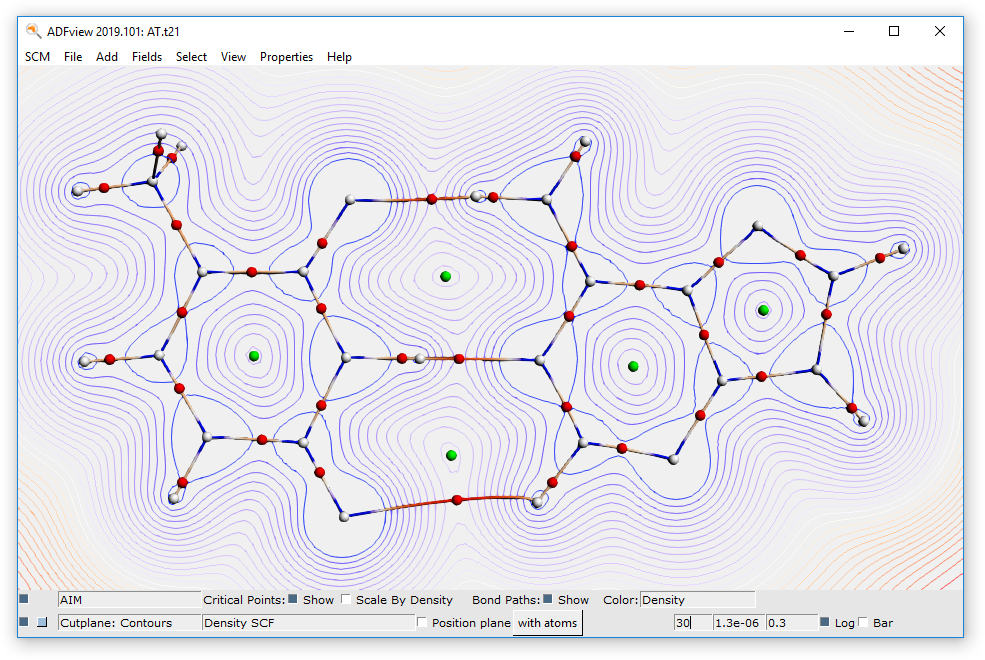
This concludes the AT QTAIM tutorial. To close all its windows:
- SCM → Quit
Benzene QTAIM charge analysis and NBOs¶
- Start ADFinputMake a benzene molecule (for example by searching for it with cmd/ctrl-F)Set up a Single Point calculation without frozen core (Frozen Core → None)Panel bar Properties → QTAIMCheck the ‘Perform QTAIM analysis’ optionSet the Analysis level to ExtendedPanel bar Properties → Localized Orbitals, NBOCheck the ‘Perform NBO analysis’ optionRequest Boys-Foster localized orbitalsRun this setup (File → Run)
When the calculation is done (it should run very fast), we use ADFview to examine the QTAIM charges and compare them with Mulliken charges:
- Open the results with ADFviewShow the QTAIM atomic charges (Properties → Atom Info → QTAIM Charge → Show)Color the atoms by QTAIM charges (Properties → Color Atoms By → QTAIM Charge)Show the Mulliken charges (Properties → Atom Info → Mulliken Charge → Show)
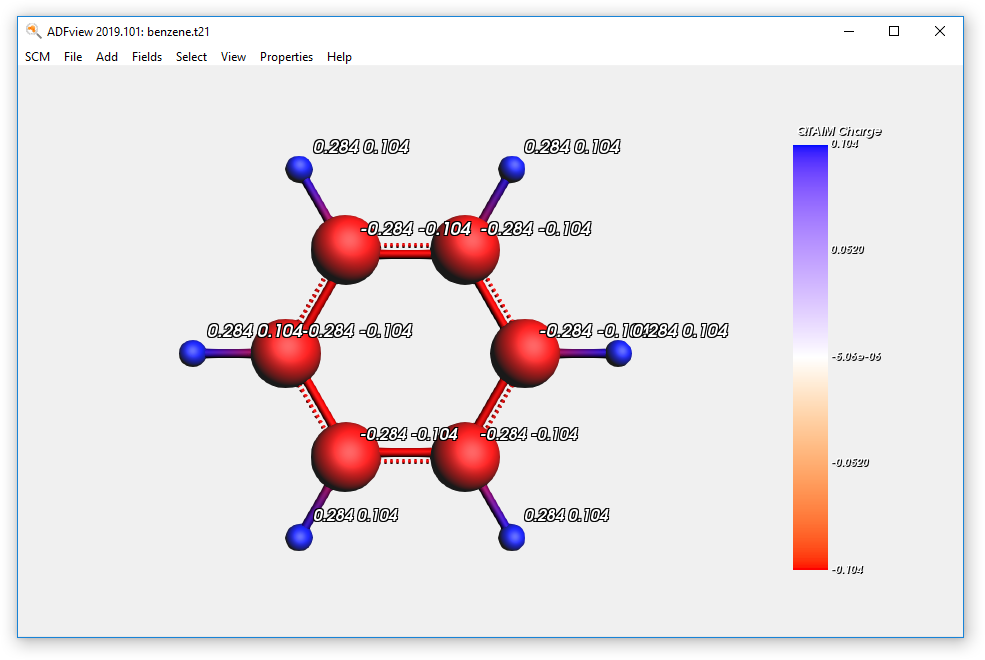
Next we inspect the NBOs and Boys-Foster localized orbitals. To remove the charge display we close and open ADFview, but you could also have used the View menu to remove them by hand:
- Close ADFviewOpen the results again with ADFviewAdd an isosurface with phaseUse the field menu in the new control line,and observe the labels present with the NBOs and NLMOsOpen a NBO similar to BD Cn - HnImprove the grid by using Fields → Grid → Fine

Obviously, you can also visualize the NLMOs or the Boys-Foster localized orbitals (which are just called Localized Orbitals in the fields menu.
Rationalizing a typical SN2 reaction using condensed Conceptual DFT descriptors¶
The chemical reactivity of reactants or key intermediates can be analyzed using condensed (over QTAIM basins) Conceptual DFT descriptors such as Fukui functions or Dual Descriptor. We strongly suggest the use of the Dual Descriptor, which gives at one glance a more complete description of reactivity behaviors. All the following calculations are based on frontier molecular orbitals (FMOs) using Koopmans approximation, which presents advantages (fast calculations) and drawbacks (in particular if FMOs are degenerated or quasi-degenerated).
An alternative way, based on finite difference linear (FDL) approximation, is available in ADF: Fukui Functions and Dual Descriptor. The FDL approximation offers a more rigorous approach, but it requires three calculations (systems with N electrons (reference), N+δ electrons and N-δ electrons (0<δ<=1)) and shows other drawbacks. For instance, adding one electron to the reference system may lead to unconverged SCF procedure, or the corresponding spin states might be unobvious. Besides, some ambiguity remains about which atomic basins (relaxed or unrelaxed) should be used when adding or removing electrons.

- Run this setup: File → Run, use ‘nucleophile’ as file name for your jobWait until it is ready, click then No when asked to update the coordinates in ADFinput
At the end of the optimization process, all the QTAIM properties will be calculated.
- Start ADFview: SCM → View
Show the condensed (over QTAIM atomic basins) ‘Fukui Fminus function’ indices that characterize the nucleophilicity of atomic sites:
- Properties → Atom Info → Fukui- (FMO) → ShowProperties → Color Atoms By → Fukui- (FMO)Properties → Atom Info → Name → Show

On this picture, we clearly see that the nitrogen site is the most nucleophilic one. To obtain a more complete picture at one glance, we can visualize the condensed values of the dual descriptor (DD) that corresponds, using the Koopmans’ theorem, to the difference between FMOs electron densities.
To this end, first hide the previous values and display the condensed DD values:
- Properties → Atom Info → Fukui- (FMO) → HideProperties → Atom Info → Dual (FMO) → ShowProperties → Color Atoms By → Dual (FMO)

Positive values correspond to atomic sites where electrophilicity is predominant, while negative values correspond to atomic sites where nucleophilicity is predominant (again, the nitrogen atom is highly nucleophilic).
In a new input window, now make the benzyl chloride (electrophile):
- SCM → New inputMake benzyl chloride by copying the following coordinates and pasting them in the ADFInput molecule drawing area:
C -0.70294970 0.03823073 0.00000000
C -0.02771734 -1.20050280 0.00000000
C 1.37040750 -1.24326069 0.00000000
C 2.10941268 -0.05859271 -0.00000000
C 1.45241936 1.17312771 -0.00000000
C 0.05527963 1.22223527 -0.00000000
C -2.21056076 0.15917615 -0.00000000
Cl -2.96962094 0.22007043 1.61845248
H -0.56397603 -2.13845972 0.00000000
H 1.88164983 -2.19732981 0.00000000
H 3.19110656 -0.09523365 -0.00000000
H 2.02573037 2.09116490 -0.00000000
H -0.43823632 2.18658642 -0.00000000
H -2.49816320 1.08415158 -0.54318756
H -2.64194499 -0.70811986 -0.54318753
- Select the Geometry Optimization taskPanel bar Properties → Conceptual DFTCheck the ‘Conceptual DFT’ optionSet the ‘Analysis level’ option to ExtendedRun this setup: File → Run, use ‘‘electrophile’’ as file name for your jobWait until it is ready, click then No when asked to update the coordinates in ADFinput
At the end of the optimization process, all the QTAIM properties will be calculated.
- Start ADFview: SCM → View
Show the condensed (over QTAIM atomic basins) ‘Fukui+ (FMO) function’ values that characterize the electrophilicity of atomic sites:
- Properties → Atom Info → Fukui+ (FMO) → ShowProperties → Color Atoms By → Fukui+ (FMO)Properties → Atom Info → Name → Show

On this picture, two carbon sites (C(4) and C(7)) have similar Fukui+ indices. Moreover, chlorine has a strong electrophilic character due to the existence of a sigma hole in the outer part of its valence shell along the C-Cl bond. Therefore, it is difficult to unambiguously determine the reactivity of this molecule by the sole QTAIM condensed Fukui+ values. In that case, the dual descriptor is quite useful, providing a balanced picture, since it allows evaluating the predominant reactivity behavior at each atomic site.
To this end, first hide the previous values and display the condensed DD values:
- Properties → Atom Info → Fukui+ (FMO) → HideProperties → Atom Info → Dual (FMO) → ShowProperties → Color Atoms By → Dual (FMO)

As already mentioned, positive values correspond to atomic sites where electrophilicity is predominant, while negative values correspond to atomic sites where nucleophilicity is predominant.
In this picture we clearly see, as expected from chemical intuition, that C(7) is highly electrophilic (compared to the other carbon atoms). This site will thus undergo a nucleophilic attack during the SN2 reaction with the N,N-dimethylbutylamine molecule, leading to the formation of a quaternary ammonium salt.
Besides, we can also observe that the chlorine atom is predominantly a nucleophilic site (due to its lone pairs) despite the presence of an electrophilic sigma hole.
- SCM→ Quit All

